Medical Image Analysis ( IF 10.9 ) Pub Date : 2022-05-25 , DOI: 10.1016/j.media.2022.102484 Alena U Uus 1 , Irina Grigorescu 1 , Milou P M van Poppel 2 , Johannes K Steinweg 2 , Thomas A Roberts 1 , Mary A Rutherford 3 , Joseph V Hajnal 4 , David F A Lloyd 2 , Kuberan Pushparajah 2 , Maria Deprez 1
|
Slice-to-volume registration (SVR) methods allow reconstruction of high-resolution 3D images from multiple motion-corrupted stacks. SVR-based pipelines have been increasingly used for motion correction for T2-weighted structural fetal MRI since they allow more informed and detailed diagnosis of brain and body anomalies including congenital heart defects (Lloyd et al., 2019). Recently, fully automated rigid SVR reconstruction of the fetal brain in the atlas space was achieved in Salehi et al. (2019) that used convolutional neural networks (CNNs) for segmentation and pose estimation. However, these CNN-based methods have not yet been applied to the fetal trunk region. Meanwhile, the existing rigid and deformable SVR (DSVR) solutions (Uus et al., 2020) for the fetal trunk region are limited by the requirement of manual input as well the narrow capture range of the classical gradient descent based registration methods that cannot resolve severe fetal motion frequently occurring at the early gestational age (GA). Furthermore, in our experience, the conventional 2D slice-wise CNN-based brain masking solutions are reportedly prone to errors that require manual corrections when applied on a wide range of acquisition protocols or abnormal cases in clinical setting.
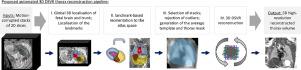
In this work, we propose a fully automated pipeline for reconstruction of the fetal thorax region for 21–36 weeks GA range T2-weighted MRI datasets. It includes 3D CNN-based intra-uterine localisation of the fetal trunk and landmark-guided pose estimation steps that allow automated DSVR reconstruction in the standard radiological space irrespective of the fetal trunk position or the regional stack coverage. The additional step for generation of the common template space and rejection of outliers provides the means for automated exclusion of stacks affected by low image quality or extreme motion. The pipeline was quantitatively evaluated on a series of experiments including fetal MRI datasets and simulated rotation motion. Furthermore, we performed a qualitative assessment of the image reconstruction quality in terms of the definition of vascular structures on 100 early (median 23.14 weeks) and late (median 31.79 weeks) GA group MRI datasets covering 21 to 36 weeks GA range.
中文翻译:

在 21-36 周的 GA 范围内,根据运动损坏的 MRI 堆栈在标准图谱空间中自动 3D 重建胎儿胸部
切片到体积配准 (SVR) 方法允许从多个运动损坏的堆栈重建高分辨率 3D 图像。基于 SVR 的管道已越来越多地用于 T2 加权结构性胎儿 MRI 的运动校正,因为它们允许对包括先天性心脏缺陷在内的大脑和身体异常进行更明智和更详细的诊断(Lloyd 等人,2019 年)。最近,Salehi 等人实现了图谱空间中胎儿大脑的全自动刚性 SVR 重建。(2019) 使用卷积神经网络 (CNN) 进行分割和姿态估计。然而,这些基于 CNN 的方法尚未应用于胎儿躯干区域。同时,现有的刚性和可变形 SVR(DSVR)解决方案(Uus 等人,2020) 的胎儿躯干区域受限于手动输入的要求以及基于经典梯度下降的配准方法的狭窄捕获范围,无法解决在早期胎龄 (GA) 经常发生的严重胎儿运动。此外,根据我们的经验,据报道,传统的基于 CNN 的二维切片脑掩蔽解决方案在应用于广泛的采集协议或临床环境中的异常情况时,容易出现需要手动更正的错误。
在这项工作中,我们提出了一个全自动管道,用于重建 21-36 周 GA 范围 T2 加权 MRI 数据集的胎儿胸部区域。它包括基于 3D CNN 的胎儿躯干宫内定位和界标引导的姿势估计步骤,这些步骤允许在标准放射学空间中进行自动 DSVR 重建,而不管胎儿躯干位置或区域堆栈覆盖范围如何。生成通用模板空间和拒绝异常值的附加步骤提供了自动排除受低图像质量或极端运动影响的堆栈的方法。该管道在一系列实验中进行了定量评估,包括胎儿 MRI 数据集和模拟旋转运动。此外,



























 京公网安备 11010802027423号
京公网安备 11010802027423号