Medical Image Analysis ( IF 10.9 ) Pub Date : 2022-05-25 , DOI: 10.1016/j.media.2022.102486 Wenqi Lu 1 , Michael Toss 2 , Muhammad Dawood 1 , Emad Rakha 2 , Nasir Rajpoot 1 , Fayyaz Minhas 1
|
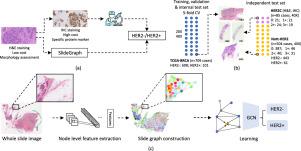
Human epidermal growth factor receptor 2 (HER2) is an important prognostic and predictive factor which is overexpressed in 15–20% of breast cancer (BCa). The determination of its status is a key clinical decision making step for selection of treatment regimen and prognostication. HER2 status is evaluated using transcriptomics or immunohistochemistry (IHC) through in-situ hybridisation (ISH) which incurs additional costs and tissue burden and is prone to analytical variabilities in terms of manual observational biases in scoring. In this study, we propose a novel graph neural network (GNN) based model (SlideGraph) to predict HER2 status directly from whole-slide images of routine Haematoxylin and Eosin (H&E) stained slides. The network was trained and tested on slides from The Cancer Genome Atlas (TCGA) in addition to two independent test datasets. We demonstrate that the proposed model outperforms the state-of-the-art methods with area under the ROC curve (AUC) values 0.75 on TCGA and 0.80 on independent test sets. Our experiments show that the proposed approach can be utilised for case triaging as well as pre-ordering diagnostic tests in a diagnostic setting. It can also be used for other weakly supervised prediction problems in computational pathology. The SlideGraph code repository is available at https://github.com/wenqi006/SlideGraph along with an IPython notebook showing an end-to-end use case at https://github.com/TissueImageAnalytics/tiatoolbox/blob/develop/examples/full-pipelines/slide-graph.ipynb.
中文翻译:

SlideGraph +:整个幻灯片图像级别图来预测乳腺癌中的 HER2 状态
人表皮生长因子受体 2 (HER2) 是一种重要的预后和预测因子,在 15-20% 的乳腺癌 (BCa) 中过度表达。确定其状态是选择治疗方案和预后的关键临床决策步骤。HER2 状态通过原位杂交 (ISH) 使用转录组学或免疫组织化学 (IHC) 进行评估,这会产生额外的成本和组织负担,并且在评分中的手动观察偏差方面容易出现分析变异性。在这项研究中,我们提出了一种新的基于图神经网络 (GNN) 的模型 (SlideGraph) 直接从常规苏木精和伊红 (H&E) 染色载玻片的全载玻片图像预测 HER2 状态。除了两个独立的测试数据集外,该网络还在癌症基因组图谱 (TCGA) 的幻灯片上进行了训练和测试。我们证明了所提出的模型在 ROC 曲线下面积 (AUC) 值方面优于最先进的方法TCGA 为 0.75,独立测试集为 0.80。我们的实验表明,所提出的方法可用于病例分类以及在诊断环境中预先订购诊断测试。它也可以用于计算病理学中的其他弱监督预测问题。幻灯片图表代码存储库可在 https://github.com/wenqi006/SlideGraph 以及显示端到端用例的 IPython 笔记本在 https://github.com/TissueImageAnalytics/tiatoolbox/blob/develop/examples/full -pipelines/slide-graph.ipynb。



























 京公网安备 11010802027423号
京公网安备 11010802027423号