当前位置:
X-MOL 学术
›
Mol. Microbiol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
The complex Tup1-Cyc8 bridges transcription factor ClrB and putative histone methyltransferase LaeA to activate the expression of cellulolytic genes
Molecular Microbiology ( IF 3.6 ) Pub Date : 2022-01-24 , DOI: 10.1111/mmi.14885 Xiujun Zhang 1, 2, 3 , Yueyan Hu 1, 2 , Guodong Liu 1, 2 , Meng Liu 2 , Zhonghai Li 2, 4 , Jian Zhao 2 , Xin Song 1, 2 , Yaohua Zhong 2 , Yinbo Qu 1, 2 , Lushan Wang 2 , Yuqi Qin 1, 2
Molecular Microbiology ( IF 3.6 ) Pub Date : 2022-01-24 , DOI: 10.1111/mmi.14885 Xiujun Zhang 1, 2, 3 , Yueyan Hu 1, 2 , Guodong Liu 1, 2 , Meng Liu 2 , Zhonghai Li 2, 4 , Jian Zhao 2 , Xin Song 1, 2 , Yaohua Zhong 2 , Yinbo Qu 1, 2 , Lushan Wang 2 , Yuqi Qin 1, 2
Affiliation
|
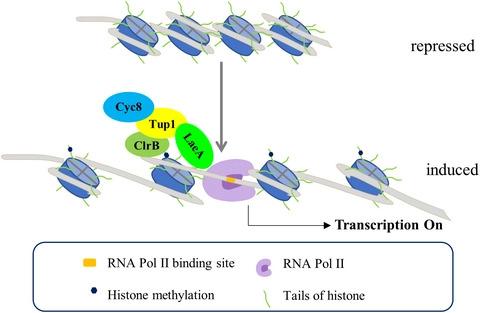
The degradation of lignocellulosic biomass by cellulolytic enzymes is involved in the global carbon cycle. The hydrolysis of lignocellulosic biomass into fermentable sugars is potential as an excellent industrial resource to produce a variety of chemical products. The production of cellulolytic enzymes is regulated mainly at the transcriptional level in filamentous fungi. Transcription factor ClrB and the putative histone methyltransferase LaeA, are both necessary for the expression of cellulolytic genes. However, the mechanism by which transcription factors and methyltransferase coordinately regulate cellulolytic genes is still unknown. Here, we reveal a transcriptional regulatory mechanism involving Penicillium oxalicum transcription factor ClrB (PoClrB), complex Tup1-Cyc8, and putative histone methyltransferase LaeA (PoLaeA). As the transcription factor, PoClrB binds the targeted promoters of cellulolytic genes, recruits PoTup1-Cyc8 complex via direct interaction with PoTup1. PoTup1 interacts with PoCyc8 to form the coactivator complex PoTup1-Cyc8. Then, PoTup1 recruits putative histone methyltransferase PoLaeA to modify the chromatin structure of the upstream region of cellulolytic genes, thereby facilitating the binding of transcription machinery to activating the corresponding cellulolytic gene expression. Our results contribute to a better understanding of complex transcriptional regulation mechanisms of cellulolytic genes and will be valuable for lignocellulosic biorefining.
中文翻译:

复杂的 Tup1-Cyc8 桥接转录因子 ClrB 和推定的组蛋白甲基转移酶 LaeA 以激活纤维素分解基因的表达
纤维素分解酶对木质纤维素生物质的降解涉及全球碳循环。木质纤维素生物质水解成可发酵糖类是生产各种化学产品的优良工业资源。纤维素分解酶的产生主要在丝状真菌的转录水平上受到调节。转录因子 ClrB 和推定的组蛋白甲基转移酶 LaeA 都是纤维素分解基因表达所必需的。然而,转录因子和甲基转移酶协同调节纤维素分解基因的机制尚不清楚。在这里,我们揭示了一种涉及草酸青霉的转录调控机制转录因子 ClrB (PoClrB)、复合物 Tup1-Cyc8 和推定的组蛋白甲基转移酶 LaeA (PoLaeA)。作为转录因子,PoClrB 结合纤维素分解基因的靶向启动子,通过与 PoTup1 的直接相互作用募集 PoTup1-Cyc8 复合物。PoTup1 与 PoCyc8 相互作用形成共激活复合物 PoTup1-Cyc8。然后,PoTup1 募集假定的组蛋白甲基转移酶 PoLaeA 来修饰纤维素分解基因上游区域的染色质结构,从而促进转录机制的结合以激活相应的纤维素分解基因表达。我们的结果有助于更好地理解纤维素分解基因的复杂转录调控机制,并将对木质纤维素生物精炼有价值。
更新日期:2022-01-24
中文翻译:

复杂的 Tup1-Cyc8 桥接转录因子 ClrB 和推定的组蛋白甲基转移酶 LaeA 以激活纤维素分解基因的表达
纤维素分解酶对木质纤维素生物质的降解涉及全球碳循环。木质纤维素生物质水解成可发酵糖类是生产各种化学产品的优良工业资源。纤维素分解酶的产生主要在丝状真菌的转录水平上受到调节。转录因子 ClrB 和推定的组蛋白甲基转移酶 LaeA 都是纤维素分解基因表达所必需的。然而,转录因子和甲基转移酶协同调节纤维素分解基因的机制尚不清楚。在这里,我们揭示了一种涉及草酸青霉的转录调控机制转录因子 ClrB (PoClrB)、复合物 Tup1-Cyc8 和推定的组蛋白甲基转移酶 LaeA (PoLaeA)。作为转录因子,PoClrB 结合纤维素分解基因的靶向启动子,通过与 PoTup1 的直接相互作用募集 PoTup1-Cyc8 复合物。PoTup1 与 PoCyc8 相互作用形成共激活复合物 PoTup1-Cyc8。然后,PoTup1 募集假定的组蛋白甲基转移酶 PoLaeA 来修饰纤维素分解基因上游区域的染色质结构,从而促进转录机制的结合以激活相应的纤维素分解基因表达。我们的结果有助于更好地理解纤维素分解基因的复杂转录调控机制,并将对木质纤维素生物精炼有价值。


























 京公网安备 11010802027423号
京公网安备 11010802027423号