Current Proteomics ( IF 0.8 ) Pub Date : 2021-09-30 , DOI: 10.2174/1570164618999201223092209 Opeyemi Iwaloye 1 , Olusola Olalekan Elekofehinti 1 , Babatomiwa Kikiowo 2 , Emmanuel Ayo Oluwarotimi 3 , Toyin Mary Fadipe 4
|
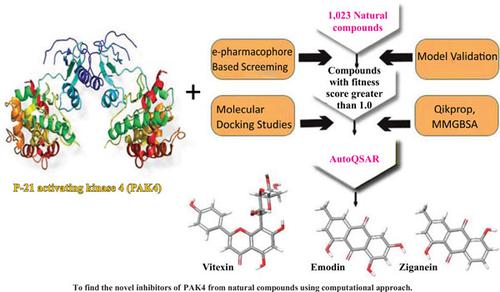
Background: P-21 activating kinase 4 (PAK4) is implicated in the poor prognosis of many cancers, especially in the progression of Triple Negative Breast Cancer (TNBC). The present study was aimed at designing some potential drug candidates as PAK4 inhibitors for breast cancer therapy.
Objective: This study aimed to finding novel inhibitors of PAK4 from natural compounds using computational approach.
Methods: An e-pharmacophore model was developed from docked PAK4-co-ligand complex and used to screen over a thousand natural compounds downloaded from BIOFACQUIM and NPASS databases to match a minimum of 5 sites for selected (ADDDHRR) hypothesis. The robustness of the virtual screening method was accessed by well-established methods including EF, ROC, BEDROC, AUAC, and the RIE. Compounds with fitness score greater than one were filtered by applying molecular docking (HTVS, SP, XP and Induced fit docking) and ADME prediction. Using Machine learning-based approach QSAR model was generated using Automated QSAR. The computed top model kpls_des_17 (R2= 0.8028, RMSE = 0.4884 and Q2 = 0.7661) was used to predict the pIC50 of the lead compounds. Internal and external validations were accessed to determine the predictive quality of the model. Finally, the binding free energy calculation was computed.
Results: The robustness/predictive quality of the models was affirmed. The hits had better binding affinity than the reference drug and interacted with key amino acids for PAK4 inhibition. Overall, the present analysis yielded three potential inhibitors that are predicted to bind with PAK4 better than the reference drug tamoxifen. The three potent novel inhibitors, vitexin, emodin and ziganein recorded IFD score of -621.97 kcal/mol, -616.31 kcal/mol and -614.95 kcal/mol, respectively while showing moderation for ADME properties and inhibition constant.
Conclusion: It is expected that the findings reported in this study may provide insight for designing effective and less toxic PAK4 inhibitors for triple negative breast cancer.
中文翻译:

基于机器学习的虚拟筛选策略揭示了一些天然化合物作为三阴性乳腺癌的潜在 PAK4 抑制剂
背景:P-21 激活激酶 4 (PAK4) 与许多癌症的不良预后有关,尤其是与三阴性乳腺癌 (TNBC) 的进展有关。本研究旨在设计一些潜在的候选药物作为 PAK4 抑制剂用于乳腺癌治疗。
目的:本研究旨在使用计算方法从天然化合物中寻找新型 PAK4 抑制剂。
方法:电子药效团模型是从对接的 PAK4 共配体复合物开发的,用于筛选从 BIOFACQUIM 和 NPASS 数据库下载的一千多种天然化合物,以匹配至少 5 个站点的选定 (ADDDHRR) 假设。虚拟筛选方法的稳健性可通过完善的方法进行评估,包括 EF、ROC、BEDROC、AUAC 和 RIE。通过应用分子对接(HTVS、SP、XP 和诱导拟合对接)和 ADME 预测过滤适合度得分大于 1 的化合物。使用基于机器学习的方法 QSAR 模型是使用自动 QSAR 生成的。计算的顶级模型 kpls_des_17(R2= 0.8028,RMSE = 0.4884 和 Q2 = 0.7661)用于预测先导化合物的 pIC50。访问内部和外部验证以确定模型的预测质量。最后,
结果:模型的稳健性/预测质量得到肯定。命中具有比参考药物更好的结合亲和力,并且与抑制 PAK4 的关键氨基酸相互作用。总体而言,本分析产生了三种潜在的抑制剂,预计它们比参考药物他莫昔芬更好地与 PAK4 结合。三种有效的新型抑制剂,牡荆素、大黄素和齐甘素的 IFD 评分分别为 -621.97 kcal/mol、-616.31 kcal/mol 和 -614.95 kcal/mol,同时显示出对 ADME 特性和抑制常数的调节。
结论:预计本研究报告的发现可为设计有效且毒性较低的三阴性乳腺癌 PAK4 抑制剂提供见解。


























 京公网安备 11010802027423号
京公网安备 11010802027423号