Environment International ( IF 11.8 ) Pub Date : 2021-11-13 , DOI: 10.1016/j.envint.2021.106978 Liping Ma 1 , Huiying Yang 1 , Lei Guan 1 , Xiaoyu Liu 1 , Tong Zhang 2
|
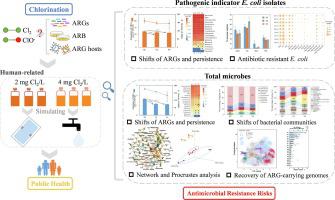
As a widely used disinfection technology, the effects of chlorination on antibiotic resistome and bacterial community received great scientific concerns, while the pathogens associated health risks kept largely unknown. With this concern, the present study used metagenomic analysis combined with culture method to reveal chlorination effects on antibiotic resistance genes (ARGs) and their bacterial hosts (total microbes and Escherichia coli) through simulating the chlorination dosage with human health concerns (drinking water and swimming pool). The resistome profiling showed that chlorination process could significantly decrease both abundance and diversity of total ARGs, while with limited removal rates of 6.0–8.7% for opportunistic pathogens E. coli isolates. Of all the observed 515 ARG subtypes, 105 core subtypes were identified and persistent during chlorination for both total microbes and E. coli. Antibiotic susceptibility test showed that chlorination treatment could efficiently remove multi-resistant E. coli isolates but select for tetracycline resistant isolates. Five ARG-carrying genomes (assigned to Bacteroidetes, Firmicutes, Actinobacteria) enriched by 18.1–102% after chlorination were retrieved by using metagenomic binning strategies. Bray-Curtis dissimilarity, network and procrustes analyses all indicated the remained antibiotic resistome and bacterial community were mainly chlorination-driven. Furthermore, a systematic pipeline for monitoring chlorination-associated antimicrobial resistance risks was proposed. These together enhance our knowledge of chlorination treatment associated public concerns, as important reference and guidance for surveillance and control of antibiotic resistance.
中文翻译:

氯化消毒下抗生素耐药基因和耐药性风险与公共卫生问题
作为一种广泛使用的消毒技术,氯化对抗生素耐药性和细菌群落的影响受到了极大的科学关注,而与健康风险相关的病原体却在很大程度上未知。鉴于此,本研究采用宏基因组分析结合培养方法,通过模拟氯化剂量与人类健康问题(饮用水和游泳)来揭示氯化对抗生素抗性基因(ARGs)及其细菌宿主(总微生物和大肠杆菌)的影响。水池)。抗性组分析表明,氯化过程可以显着降低总 ARGs 的丰度和多样性,而机会性病原体大肠杆菌的去除率有限,为 6.0-8.7%隔离。在所有观察到的 515 种 ARG 亚型中,105 种核心亚型在总微生物和大肠杆菌的氯化过程中被鉴定并持续存在。抗生素药敏试验表明氯化处理能有效去除多重耐药大肠杆菌分离株,但选择耐四环素的分离株。通过使用宏基因组分箱策略,在氯化后富集了 18.1-102% 的五个携带 ARG 的基因组(分配给拟杆菌门、厚壁菌门、放线菌门)。Bray-Curtis 相异性、网络和 procrustes 分析均表明剩余的抗生素抗性组和细菌群落主要是氯化驱动的。此外,还提出了一种监测氯化相关抗菌素耐药性风险的系统管道。这些共同提高了我们对氯化处理相关公众关注的认识,作为监测和控制抗生素耐药性的重要参考和指导。



























 京公网安备 11010802027423号
京公网安备 11010802027423号