Medical Image Analysis ( IF 10.9 ) Pub Date : 2021-11-13 , DOI: 10.1016/j.media.2021.102297 Mansu Kim 1 , Eun Jeong Min 2 , Kefei Liu 3 , Jingwen Yan 4 , Andrew J Saykin 5 , Jason H Moore 3 , Qi Long 3 , Li Shen 3
|
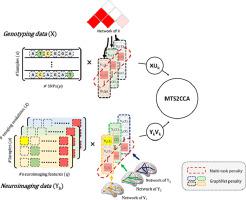
The advances in technologies for acquiring brain imaging and high-throughput genetic data allow the researcher to access a large amount of multi-modal data. Although the sparse canonical correlation analysis is a powerful bi-multivariate association analysis technique for feature selection, we are still facing major challenges in integrating multi-modal imaging genetic data and yielding biologically meaningful interpretation of imaging genetic findings. In this study, we propose a novel multi-task learning based structured sparse canonical correlation analysis (MTS2CCA) to deliver interpretable results and improve integration in imaging genetics studies. We perform comparative studies with state-of-the-art competing methods on both simulation and real imaging genetic data. On the simulation data, our proposed model has achieved the best performance in terms of canonical correlation coefficients, estimation accuracy, and feature selection accuracy. On the real imaging genetic data, our proposed model has revealed promising features of single-nucleotide polymorphisms and brain regions related to sleep. The identified features can be used to improve clinical score prediction using promising imaging genetic biomarkers. An interesting future direction is to apply our model to additional neurological or psychiatric cohorts such as patients with Alzheimer’s or Parkinson’s disease to demonstrate the generalizability of our method.
中文翻译:

基于多任务学习的脑成像遗传学结构化稀疏典型相关分析
获取脑成像和高通量遗传数据技术的进步使研究人员能够访问大量多模式数据。尽管稀疏典型相关分析是一种用于特征选择的强大的双多元关联分析技术,但我们在整合多模态成像遗传数据和对成像遗传发现产生具有生物学意义的解释方面仍然面临着重大挑战。在这项研究中,我们提出了一种新颖的基于多任务学习的结构化稀疏典型相关分析(MTS2CCA),以提供可解释的结果并改善成像遗传学研究的整合。我们使用最先进的竞争方法对模拟和真实成像遗传数据进行比较研究。在模拟数据上,我们提出的模型在典型相关系数、估计精度和特征选择精度方面取得了最佳性能。在真实的成像遗传数据上,我们提出的模型揭示了单核苷酸多态性和与睡眠相关的大脑区域的有希望的特征。所识别的特征可用于使用有前景的成像遗传生物标志物来改善临床评分预测。未来一个有趣的方向是将我们的模型应用于其他神经或精神病学队列,例如阿尔茨海默病或帕金森病患者,以证明我们方法的普遍性。

























 京公网安备 11010802027423号
京公网安备 11010802027423号