Cell ( IF 64.5 ) Pub Date : 2021-10-07 , DOI: 10.1016/j.cell.2021.09.016 Benedikt W Bauer 1 , Iain F Davidson 1 , Daniel Canena 2 , Gordana Wutz 1 , Wen Tang 1 , Gabriele Litos 1 , Sabrina Horn 3 , Peter Hinterdorfer 2 , Jan-Michael Peters 1
|
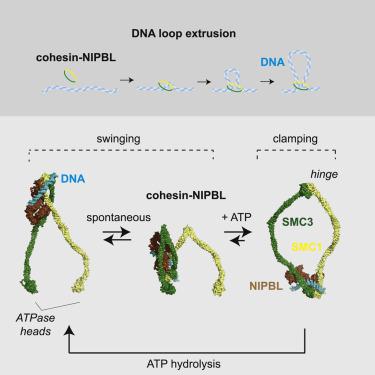
Structural maintenance of chromosomes (SMC) complexes organize genome topology in all kingdoms of life and have been proposed to perform this function by DNA loop extrusion. How this process works is unknown. Here, we have analyzed how loop extrusion is mediated by human cohesin-NIPBL complexes, which enable chromatin folding in interphase cells. We have identified DNA binding sites and large-scale conformational changes that are required for loop extrusion and have determined how these are coordinated. Our results suggest that DNA is translocated by a spontaneous 50 nm-swing of cohesin’s hinge, which hands DNA over to the ATPase head of SMC3, where upon binding of ATP, DNA is clamped by NIPBL. During this process, NIPBL “jumps ship” from the hinge toward the SMC3 head and might thereby couple the spontaneous hinge swing to ATP-dependent DNA clamping. These results reveal mechanistic principles of how cohesin-NIPBL and possibly other SMC complexes mediate loop extrusion.
中文翻译:

Cohesin 通过“摆动和夹紧”机制介导 DNA 环挤出
染色体结构维持 (SMC) 复合物在所有生命王国中组织基因组拓扑结构,并已被提议通过 DNA 环挤出来执行此功能。这个过程如何运作是未知的。在这里,我们分析了环挤出是如何由人类 cohesin-NIPBL 复合物介导的,这使得间期细胞中的染色质折叠。我们已经确定了环挤出所需的 DNA 结合位点和大规模构象变化,并确定了这些是如何协调的。我们的结果表明 DNA 通过自发的 50 nm 摆动的 cohesin 铰链转移,将 DNA 交给 SMC3 的 ATPase 头部,在那里结合 ATP,DNA 被 NIPBL 夹住。在这个过程中,NIPBL 从铰链“跳跃”到 SMC3 头部,从而可能将自发的铰链摆动与 ATP 依赖性 DNA 夹紧结合起来。这些结果揭示了 cohesin-NIPBL 和可能的其他 SMC 复合物如何介导环挤出的机械原理。



























 京公网安备 11010802027423号
京公网安备 11010802027423号