当前位置:
X-MOL 学术
›
J. Chem. Inf. Model.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Molecular Dynamics Simulations of the Aptamer Domain of Guanidinium Ion Binding Riboswitch ykkC-III: Structural Insights into the Discrimination of Cognate and Alternate Ligands
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2021-10-05 , DOI: 10.1021/acs.jcim.1c01022 Indu Negi 1 , Amanpreet Singh Mahmi 1 , Preethi Seelam Prabhakar 2 , Purshotam Sharma 1
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2021-10-05 , DOI: 10.1021/acs.jcim.1c01022 Indu Negi 1 , Amanpreet Singh Mahmi 1 , Preethi Seelam Prabhakar 2 , Purshotam Sharma 1
Affiliation
|
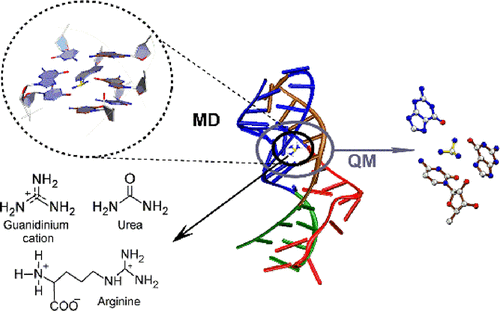
Guanidinium ion is a toxic cellular metabolite. The ykkC-III riboswitch, an mRNA stretch, regulates the gene expression by undergoing a conformational change in response to the binding of a free guanidinium ion and thereby plays a potentially important role in alleviating guanidinium toxicity in cells. An experimental crystal structure of the guanidinium-bound aptamer domain of the riboswitch from Thermobifida Fusca revealed the overall RNA architecture and mapped the specific noncovalent interactions that stabilize the ligand within the binding pocket aptamer. However, details of how the aptamer domain discriminates the cognate ligand from its closest structurally analogous physiological metabolites (arginine and urea), and how the binding of cognate ligand arrays information from the aptamer domain to the expression platform for regulating the gene expression, are not well understood. To fill this void, we perform a cumulative of 2 μs all-atom explicit-solvent molecular dynamics (MD) simulations on the full aptamer domain, augmented with quantum-chemical calculations on the ligand-binding pocket, to compare the structural and dynamical details of the guanidinium-bound state with the arginine or urea bound states, as well as the unbound (open) state. Analysis of the ligand-binding pocket reveals that due to unfavorable interactions with the binding-pocket residues, urea cannot bind the aptamer domain and thereby cannot alter the gene expression. Although interaction of the guanidyl moiety of arginine within the binding pocket is either comparable or stronger than the guanidinium ion, additional non-native hydrogen-bonding networks, as well as differences in the dynamical details of the arginine-bound state, explain why arginine cannot transmit the information from the aptamer domain to the expression platform. Based on our simulations, we propose a mechanism of how the aptamer domain communicates with the expression platform. Overall, our work provides interesting insights into the ligand recognition by a specific class of riboswitches and may hopefully inspire future studies to further understand the gene regulation by riboswitches.
中文翻译:

胍离子结合核糖开关 ykkC-III 适体结构域的分子动力学模拟:对同源和替代配体鉴别的结构洞察
胍离子是一种有毒的细胞代谢物。所述ykkC -III核糖开关中,mRNA拉伸,通过经历响应于自由胍离子的结合,从而起着在减轻细胞毒性胍潜在的重要作用的构象变化调节基因表达。来自Thermobifida Fusca的核糖开关的胍结合适体结构域的实验晶体结构揭示了整体的 RNA 结构并绘制了特定的非共价相互作用图,这些相互作用稳定了结合口袋适体内的配体。然而,适配体结构域如何将同源配体与其最接近的结构类似的生理代谢物(精氨酸和尿素)区分开来的细节,以及同源配体如何结合从适配体结构域到表达平台以调节基因表达的信息,都不是完全了解。为了填补这一空白,我们对完整的适体域进行了 2 μs 的累积全原子显式溶剂分子动力学 (MD) 模拟,并在配体结合口袋上进行了量子化学计算,以比较结构和动力学细节胍结合状态与精氨酸或尿素结合状态,以及未结合(开放)状态。配体结合袋的分析表明,由于与结合袋残基的不利相互作用,尿素不能结合适体结构域,因此不能改变基因表达。尽管结合口袋内精氨酸的胍基部分的相互作用与胍离子相当或更强,但额外的非天然氢键网络,以及精氨酸结合状态的动力学细节的差异,解释了为什么精氨酸不能将信息从适体域传输到表达平台。基于我们的模拟,我们提出了适体域如何与表达平台通信的机制。总体,
更新日期:2021-10-25
中文翻译:

胍离子结合核糖开关 ykkC-III 适体结构域的分子动力学模拟:对同源和替代配体鉴别的结构洞察
胍离子是一种有毒的细胞代谢物。所述ykkC -III核糖开关中,mRNA拉伸,通过经历响应于自由胍离子的结合,从而起着在减轻细胞毒性胍潜在的重要作用的构象变化调节基因表达。来自Thermobifida Fusca的核糖开关的胍结合适体结构域的实验晶体结构揭示了整体的 RNA 结构并绘制了特定的非共价相互作用图,这些相互作用稳定了结合口袋适体内的配体。然而,适配体结构域如何将同源配体与其最接近的结构类似的生理代谢物(精氨酸和尿素)区分开来的细节,以及同源配体如何结合从适配体结构域到表达平台以调节基因表达的信息,都不是完全了解。为了填补这一空白,我们对完整的适体域进行了 2 μs 的累积全原子显式溶剂分子动力学 (MD) 模拟,并在配体结合口袋上进行了量子化学计算,以比较结构和动力学细节胍结合状态与精氨酸或尿素结合状态,以及未结合(开放)状态。配体结合袋的分析表明,由于与结合袋残基的不利相互作用,尿素不能结合适体结构域,因此不能改变基因表达。尽管结合口袋内精氨酸的胍基部分的相互作用与胍离子相当或更强,但额外的非天然氢键网络,以及精氨酸结合状态的动力学细节的差异,解释了为什么精氨酸不能将信息从适体域传输到表达平台。基于我们的模拟,我们提出了适体域如何与表达平台通信的机制。总体,


























 京公网安备 11010802027423号
京公网安备 11010802027423号