当前位置:
X-MOL 学术
›
Ecol. Lett.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Reconstructing large interaction networks from empirical time series data
Ecology Letters ( IF 8.8 ) Pub Date : 2021-10-03 , DOI: 10.1111/ele.13897 Chun-Wei Chang, Takeshi Miki, Masayuki Ushio, Po-Ju Ke, Hsiao-Pei Lu, Fuh-Kwo Shiah, Chih-hao Hsieh
Ecology Letters ( IF 8.8 ) Pub Date : 2021-10-03 , DOI: 10.1111/ele.13897 Chun-Wei Chang, Takeshi Miki, Masayuki Ushio, Po-Ju Ke, Hsiao-Pei Lu, Fuh-Kwo Shiah, Chih-hao Hsieh
|
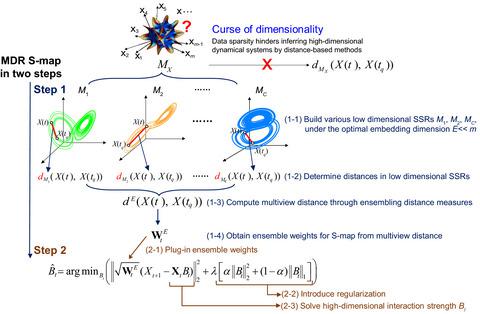
Reconstructing interactions from observational data is a critical need for investigating natural biological networks, wherein network dimensionality is usually high. However, these pose a challenge to existing methods that can quantify only small interaction networks. Here, we proposed a novel approach to reconstruct high-dimensional interaction Jacobian networks using empirical time series without specific model assumptions. This method, named “multiview distance regularised S-map,” generalised the state space reconstruction to accommodate high dimensionality and overcome difficulties in quantifying massive interactions with limited data. When evaluating this method using time series generated from theoretical models involving hundreds of interacting species, estimated strengths of interaction Jacobians were in good agreement with theoretical expectations. Applying this method to a natural bacterial community helped identify important species from the interaction network and revealed mechanisms governing the dynamical stability of a bacterial community. The proposed method overcame the challenge of high dimensionality in large natural dynamical systems.
中文翻译:

从经验时间序列数据重建大型交互网络
从观测数据重建相互作用是研究自然生物网络的关键需求,其中网络维数通常很高。然而,这些对只能量化小型交互网络的现有方法构成了挑战。在这里,我们提出了一种使用经验时间序列重建高维交互雅可比网络的新方法,无需特定的模型假设。这种名为“多视角距离正则化 S-map”的方法概括了状态空间重建以适应高维,并克服了用有限数据量化大规模交互的困难。当使用从涉及数百个相互作用物种的理论模型生成的时间序列评估此方法时,相互作用雅可比矩阵的估计强度与理论预期非常一致。将此方法应用于天然细菌群落有助于从相互作用网络中识别重要物种,并揭示控制细菌群落动态稳定性的机制。所提出的方法克服了大型自然动力系统中高维的挑战。
更新日期:2021-11-11
中文翻译:

从经验时间序列数据重建大型交互网络
从观测数据重建相互作用是研究自然生物网络的关键需求,其中网络维数通常很高。然而,这些对只能量化小型交互网络的现有方法构成了挑战。在这里,我们提出了一种使用经验时间序列重建高维交互雅可比网络的新方法,无需特定的模型假设。这种名为“多视角距离正则化 S-map”的方法概括了状态空间重建以适应高维,并克服了用有限数据量化大规模交互的困难。当使用从涉及数百个相互作用物种的理论模型生成的时间序列评估此方法时,相互作用雅可比矩阵的估计强度与理论预期非常一致。将此方法应用于天然细菌群落有助于从相互作用网络中识别重要物种,并揭示控制细菌群落动态稳定性的机制。所提出的方法克服了大型自然动力系统中高维的挑战。


























 京公网安备 11010802027423号
京公网安备 11010802027423号