当前位置:
X-MOL 学术
›
J. Chem. Inf. Model.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
EHreact: Extended Hasse Diagrams for the Extraction and Scoring of Enzymatic Reaction Templates
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2021-09-29 , DOI: 10.1021/acs.jcim.1c00921 Esther Heid 1 , Samuel Goldman 2 , Karthik Sankaranarayanan 1 , Connor W Coley 1 , Christoph Flamm 3 , William H Green 1
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2021-09-29 , DOI: 10.1021/acs.jcim.1c00921 Esther Heid 1 , Samuel Goldman 2 , Karthik Sankaranarayanan 1 , Connor W Coley 1 , Christoph Flamm 3 , William H Green 1
Affiliation
|
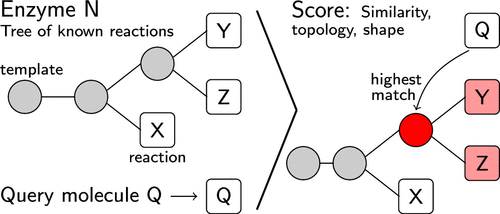
Data-driven computer-aided synthesis planning utilizing organic or biocatalyzed reactions from large databases has gained increasing interest in the last decade, sparking the development of numerous tools to extract, apply, and score general reaction templates. The generation of reaction rules for enzymatic reactions is especially challenging since substrate promiscuity varies between enzymes, causing the optimal levels of rule specificity and optimal number of included atoms to differ between enzymes. This complicates an automated extraction from databases and has promoted the creation of manually curated reaction rule sets. Here, we present EHreact, a purely data-driven open-source software tool, to extract and score reaction rules from sets of reactions known to be catalyzed by an enzyme at appropriate levels of specificity without expert knowledge. EHreact extracts and groups reaction rules into tree-like structures, Hasse diagrams, based on common substructures in the imaginary transition structures. Each diagram can be utilized to output a single or a set of reaction rules, as well as calculate the probability of a new substrate to be processed by the given enzyme by inferring information about the reactive site of the enzyme from the known reactions and their grouping in the template tree. EHreact heuristically predicts the activity of a given enzyme on a new substrate, outperforming current approaches in accuracy and functionality.
中文翻译:

EHreact:用于酶促反应模板提取和评分的扩展哈斯图
在过去十年中,利用来自大型数据库的有机或生物催化反应的数据驱动的计算机辅助合成计划引起了越来越多的兴趣,从而引发了许多用于提取、应用和评分通用反应模板的工具的开发。酶促反应的反应规则的生成尤其具有挑战性,因为酶之间的底物混杂程度不同,导致规则特异性的最佳水平和所含原子的最佳数量因酶而异。这使从数据库中自动提取变得复杂,并促进了手动策划的反应规则集的创建。在这里,我们介绍 EHreact,一个纯数据驱动的开源软件工具,在没有专业知识的情况下,从已知由酶催化的反应组中提取反应规则并以适当的特异性水平进行评分。EHreact 根据想象中的过渡结构中的常见子结构,将反应规则提取并分组为树状结构,即哈斯图。每个图表可用于输出单个或一组反应规则,以及通过从已知反应及其分组推断有关酶的反应位点的信息来计算给定酶处理新底物的概率在模板树中。EHreact 启发式地预测给定酶在新底物上的活性,在准确性和功能性方面优于当前方法。基于假想过渡结构中的公共子结构。每个图表可用于输出单个或一组反应规则,以及通过从已知反应及其分组推断有关酶的反应位点的信息来计算给定酶处理新底物的概率在模板树中。EHreact 启发式地预测给定酶在新底物上的活性,在准确性和功能性方面优于当前方法。基于假想过渡结构中的公共子结构。每个图表可用于输出单个或一组反应规则,以及通过从已知反应及其分组推断有关酶的反应位点的信息来计算给定酶处理新底物的概率在模板树中。EHreact 启发式地预测给定酶在新底物上的活性,在准确性和功能性方面优于当前方法。以及通过从已知反应及其在模板树中的分组推断有关酶的反应位点的信息,计算给定酶处理新底物的概率。EHreact 启发式地预测给定酶在新底物上的活性,在准确性和功能性方面优于当前方法。以及通过从已知反应及其在模板树中的分组推断有关酶的反应位点的信息,计算给定酶处理新底物的概率。EHreact 启发式地预测给定酶在新底物上的活性,在准确性和功能性方面优于当前方法。
更新日期:2021-10-25
中文翻译:

EHreact:用于酶促反应模板提取和评分的扩展哈斯图
在过去十年中,利用来自大型数据库的有机或生物催化反应的数据驱动的计算机辅助合成计划引起了越来越多的兴趣,从而引发了许多用于提取、应用和评分通用反应模板的工具的开发。酶促反应的反应规则的生成尤其具有挑战性,因为酶之间的底物混杂程度不同,导致规则特异性的最佳水平和所含原子的最佳数量因酶而异。这使从数据库中自动提取变得复杂,并促进了手动策划的反应规则集的创建。在这里,我们介绍 EHreact,一个纯数据驱动的开源软件工具,在没有专业知识的情况下,从已知由酶催化的反应组中提取反应规则并以适当的特异性水平进行评分。EHreact 根据想象中的过渡结构中的常见子结构,将反应规则提取并分组为树状结构,即哈斯图。每个图表可用于输出单个或一组反应规则,以及通过从已知反应及其分组推断有关酶的反应位点的信息来计算给定酶处理新底物的概率在模板树中。EHreact 启发式地预测给定酶在新底物上的活性,在准确性和功能性方面优于当前方法。基于假想过渡结构中的公共子结构。每个图表可用于输出单个或一组反应规则,以及通过从已知反应及其分组推断有关酶的反应位点的信息来计算给定酶处理新底物的概率在模板树中。EHreact 启发式地预测给定酶在新底物上的活性,在准确性和功能性方面优于当前方法。基于假想过渡结构中的公共子结构。每个图表可用于输出单个或一组反应规则,以及通过从已知反应及其分组推断有关酶的反应位点的信息来计算给定酶处理新底物的概率在模板树中。EHreact 启发式地预测给定酶在新底物上的活性,在准确性和功能性方面优于当前方法。以及通过从已知反应及其在模板树中的分组推断有关酶的反应位点的信息,计算给定酶处理新底物的概率。EHreact 启发式地预测给定酶在新底物上的活性,在准确性和功能性方面优于当前方法。以及通过从已知反应及其在模板树中的分组推断有关酶的反应位点的信息,计算给定酶处理新底物的概率。EHreact 启发式地预测给定酶在新底物上的活性,在准确性和功能性方面优于当前方法。



























 京公网安备 11010802027423号
京公网安备 11010802027423号