当前位置:
X-MOL 学术
›
J. Chem. Inf. Model.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
AlphaFold 2: Why It Works and Its Implications for Understanding the Relationships of Protein Sequence, Structure, and Function
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2021-09-29 , DOI: 10.1021/acs.jcim.1c01114 Jeffrey Skolnick 1 , Mu Gao 1 , Hongyi Zhou 1 , Suresh Singh 2
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2021-09-29 , DOI: 10.1021/acs.jcim.1c01114 Jeffrey Skolnick 1 , Mu Gao 1 , Hongyi Zhou 1 , Suresh Singh 2
Affiliation
|
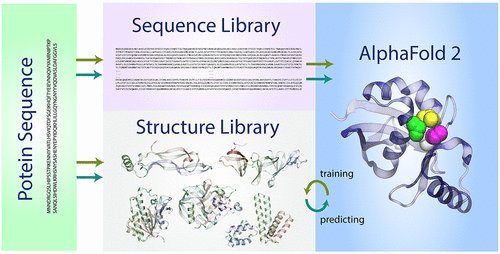
AlphaFold 2 (AF2) was the star of CASP14, the last biannual structure prediction experiment. Using novel deep learning, AF2 predicted the structures of many difficult protein targets at or near experimental resolution. Here, we present our perspective of why AF2 works and show that it is a very sophisticated fold recognition algorithm that exploits the completeness of the library of single domain PDB structures. It has also learned local side chain packing rearrangements that enable it to refine proteins to high resolution. The benefits and limitations of its ability to predict the structures of many more proteins at or close to atomic detail are discussed.
中文翻译:

AlphaFold 2:它的作用及其对理解蛋白质序列、结构和功能关系的影响
AlphaFold 2 (AF2) 是 CASP14 的明星,这是最后一个两年一次的结构预测实验。使用新的深度学习,AF2 以实验分辨率或接近实验分辨率预测了许多困难蛋白质目标的结构。在这里,我们展示了我们对 AF2 为何起作用的观点,并表明它是一种非常复杂的折叠识别算法,它利用了单域 PDB 结构库的完整性。它还学习了局部侧链堆积重排,使其能够将蛋白质提炼到高分辨率。讨论了其在或接近原子细节处预测更多蛋白质结构的能力的优点和局限性。
更新日期:2021-10-25
中文翻译:

AlphaFold 2:它的作用及其对理解蛋白质序列、结构和功能关系的影响
AlphaFold 2 (AF2) 是 CASP14 的明星,这是最后一个两年一次的结构预测实验。使用新的深度学习,AF2 以实验分辨率或接近实验分辨率预测了许多困难蛋白质目标的结构。在这里,我们展示了我们对 AF2 为何起作用的观点,并表明它是一种非常复杂的折叠识别算法,它利用了单域 PDB 结构库的完整性。它还学习了局部侧链堆积重排,使其能够将蛋白质提炼到高分辨率。讨论了其在或接近原子细节处预测更多蛋白质结构的能力的优点和局限性。



























 京公网安备 11010802027423号
京公网安备 11010802027423号