当前位置:
X-MOL 学术
›
Environ. Sci. Technol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Identification of Antimonate Reducing Bacteria and Their Potential Metabolic Traits by the Combination of Stable Isotope Probing and Metagenomic-Pangenomic Analysis
Environmental Science & Technology ( IF 11.4 ) Pub Date : 2021-09-28 , DOI: 10.1021/acs.est.1c03967 Weimin Sun 1, 2 , Xiaoxu Sun 1, 2 , Max M Häggblom 3 , Max Kolton 1, 2 , Ling Lan 1, 2 , Baoqin Li 1, 2 , Yiran Dong 4 , Rui Xu 1, 2 , Fangbai Li 1, 2
Environmental Science & Technology ( IF 11.4 ) Pub Date : 2021-09-28 , DOI: 10.1021/acs.est.1c03967 Weimin Sun 1, 2 , Xiaoxu Sun 1, 2 , Max M Häggblom 3 , Max Kolton 1, 2 , Ling Lan 1, 2 , Baoqin Li 1, 2 , Yiran Dong 4 , Rui Xu 1, 2 , Fangbai Li 1, 2
Affiliation
|
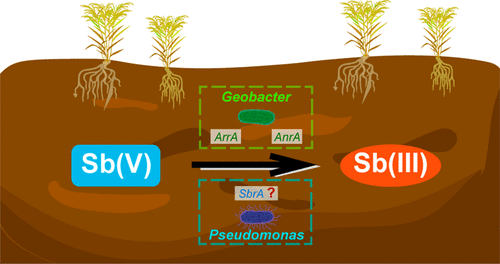
Microorganisms play an important role in altering antimony (Sb) speciation, mobility, and bioavailability, but the understanding of the microorganisms responsible for Sb(V) reduction has been limited. In this study, DNA-stable isotope probing (DNA-SIP) and metagenomics analysis were combined to identify potential Sb(V)-reducing bacteria (SbRB) and predict their metabolic pathways for Sb(V) reduction. Soil slurry cultures inoculated with Sb-contaminated paddy soils from two Sb-contaminated sites demonstrated the capability to reduce Sb(V). DNA-SIP identified bacteria belonging to the genera Pseudomonas and Geobacter as putative SbRB in these two Sb-contaminated sites. In addition, bacteria such as Lysinibacillus and Dechloromonas may potentially participate in Sb(V) reduction. Nearly complete draft genomes of putative SbRB (i.e., Pseudomonas and Geobacter) were obtained, and the genes potentially responsible for arsenic (As) and Sb reduction (i.e., respiratory arsenate reductase (arrA) and antimonate reductase (anrA)) were examined. Notably, bins affiliated with Geobacter contained arrA and anrA genes, supporting our hypothesis that they are putative SbRB. Further, pangenomic analysis indicated that various Geobacter-associated genomes obtained from diverse habitats also contained arrA and anrA genes. In contrast, Pseudomonas may use a predicted DMSO reductase closely related to sbrA (Sb(V) reductase gene) clade II to reduce Sb(V), which may need further experiments to verify. This current work represents a demonstration of using DNA-SIP and metagenomic-binning to identify SbRB and their key genes involved in Sb(V) reduction and provides valuable data sets to link bacterial identities with Sb(V) reduction.
中文翻译:

结合稳定同位素探测和宏基因组-泛基因组分析鉴定锑酸盐还原菌及其潜在代谢性状
微生物在改变锑 (Sb) 形态、迁移率和生物利用度方面发挥着重要作用,但对负责 Sb(V) 还原的微生物的了解有限。在这项研究中,DNA 稳定同位素探测 (DNA-SIP) 和宏基因组学分析相结合,以识别潜在的 Sb(V) 还原菌 (SbRB) 并预测它们的 Sb(V) 还原代谢途径。用来自两个 Sb 污染地点的 Sb 污染稻田接种的土壤泥浆培养物证明了减少 Sb(V) 的能力。DNA-SIP在这两个 Sb 污染位点将属于假单胞菌属和地杆菌属的细菌鉴定为推定的 SbRB。此外,赖氨酸杆菌和脱氯单胞菌等细菌可能参与 Sb(V) 还原。获得了推定的 SbRB(即假单胞菌和地杆菌)的几乎完整的基因组草图,并检查了可能导致砷 (As) 和 Sb 减少的基因(即呼吸性砷酸还原酶 ( arrA ) 和锑酸还原酶 ( anrA ))。值得注意的是,与Geobacter 相关的bin包含arrA和anrA基因,支持我们的假设,即它们是推定的 SbRB。此外,泛基因组分析表明,从不同栖息地获得的各种地杆菌相关基因组也含有arrA和anrA基因。相比之下,假单胞菌可能会使用与sbrA(Sb(V) 还原酶基因)进化枝 II密切相关的预测 DMSO 还原酶来还原 Sb(V),这可能需要进一步的实验来验证。目前的工作证明了使用 DNA-SIP 和宏基因组分箱来识别 SbRB 及其参与 Sb(V) 还原的关键基因,并提供有价值的数据集将细菌身份与 Sb(V) 还原联系起来。
更新日期:2021-10-19
中文翻译:

结合稳定同位素探测和宏基因组-泛基因组分析鉴定锑酸盐还原菌及其潜在代谢性状
微生物在改变锑 (Sb) 形态、迁移率和生物利用度方面发挥着重要作用,但对负责 Sb(V) 还原的微生物的了解有限。在这项研究中,DNA 稳定同位素探测 (DNA-SIP) 和宏基因组学分析相结合,以识别潜在的 Sb(V) 还原菌 (SbRB) 并预测它们的 Sb(V) 还原代谢途径。用来自两个 Sb 污染地点的 Sb 污染稻田接种的土壤泥浆培养物证明了减少 Sb(V) 的能力。DNA-SIP在这两个 Sb 污染位点将属于假单胞菌属和地杆菌属的细菌鉴定为推定的 SbRB。此外,赖氨酸杆菌和脱氯单胞菌等细菌可能参与 Sb(V) 还原。获得了推定的 SbRB(即假单胞菌和地杆菌)的几乎完整的基因组草图,并检查了可能导致砷 (As) 和 Sb 减少的基因(即呼吸性砷酸还原酶 ( arrA ) 和锑酸还原酶 ( anrA ))。值得注意的是,与Geobacter 相关的bin包含arrA和anrA基因,支持我们的假设,即它们是推定的 SbRB。此外,泛基因组分析表明,从不同栖息地获得的各种地杆菌相关基因组也含有arrA和anrA基因。相比之下,假单胞菌可能会使用与sbrA(Sb(V) 还原酶基因)进化枝 II密切相关的预测 DMSO 还原酶来还原 Sb(V),这可能需要进一步的实验来验证。目前的工作证明了使用 DNA-SIP 和宏基因组分箱来识别 SbRB 及其参与 Sb(V) 还原的关键基因,并提供有价值的数据集将细菌身份与 Sb(V) 还原联系起来。



























 京公网安备 11010802027423号
京公网安备 11010802027423号