Journal of Advanced Research ( IF 10.7 ) Pub Date : 2021-09-28 , DOI: 10.1016/j.jare.2021.09.008 Jingcheng Zhang 1 , Nan Shen 1 , Chuang Li 1 , Xingjie Xiang 1 , Gaolei Liu 1 , Ying Gui 1 , Sean Patev 2 , David S Hibbett 2 , Kerrie Barry 3 , William Andreopoulos 3 , Anna Lipzen 3 , Robert Riley 3 , Guifen He 3 , Mi Yan 3 , Igor V Grigoriev 3, 4 , Hoi Shan Kwan 5 , Man Kit Cheung 5 , Yinbing Bian 1 , Yang Xiao 1
|
Introduction
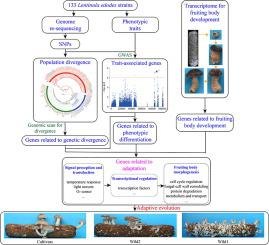
Mushroom-forming fungi comprise diverse species that develop complex multicellular structures. In cultivated species, both ecological adaptation and artificial selection have driven genome evolution. However, little is known about the connections among genotype, phenotype and adaptation in mushroom-forming fungi.
Objectives
This study aimed to (1) uncover the population structure and demographic history of Lentinula edodes, (2) dissect the genetic basis of adaptive evolution in L. edodes, and (3) determine if genes related to fruiting body development are involved in adaptive evolution.
Methods
We analyzed genomes and fruiting body-related traits (FBRTs) in 133 L. edodes strains and conducted RNA-seq analysis of fruiting body development in the YS69 strain. Combined methods of genomic scan for divergence, genome-wide association studies (GWAS), and RNA-seq were used to dissect the genetic basis of adaptive evolution.
Results
We detected three distinct subgroups of L. edodes via single nucleotide polymorphisms, which showed robust phenotypic and temperature response differentiation and correlation with geographical distribution. Demographic history inference suggests that the subgroups diverged 36,871 generations ago. Moreover, L. edodes cultivars in China may have originated from the vicinity of Northeast China. A total of 942 genes were found to be related to genetic divergence by genomic scan, and 719 genes were identified to be candidates underlying FBRTs by GWAS. Integrating results of genomic scan and GWAS, 80 genes were detected to be related to phenotypic differentiation. A total of 364 genes related to fruiting body development were involved in genetic divergence and phenotypic differentiation.
Conclusion
Adaptation to the local environment, especially temperature, triggered genetic divergence and phenotypic differentiation of L. edodes. A general model for genetic divergence and phenotypic differentiation during adaptive evolution in L. edodes, which involves in signal perception and transduction, transcriptional regulation, and fruiting body morphogenesis, was also integrated here.



























 京公网安备 11010802027423号
京公网安备 11010802027423号