当前位置:
X-MOL 学术
›
ACS Synth. Biol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Visualized Multigene Editing System for Aspergillus niger
ACS Synthetic Biology ( IF 4.7 ) Pub Date : 2021-09-24 , DOI: 10.1021/acssynbio.1c00231 Cen Li 1, 2, 3 , Jingwen Zhou 1, 2, 3, 4 , Shengqi Rao 5 , Guocheng Du 1, 2, 3 , Song Liu 1, 2, 3
ACS Synthetic Biology ( IF 4.7 ) Pub Date : 2021-09-24 , DOI: 10.1021/acssynbio.1c00231 Cen Li 1, 2, 3 , Jingwen Zhou 1, 2, 3, 4 , Shengqi Rao 5 , Guocheng Du 1, 2, 3 , Song Liu 1, 2, 3
Affiliation
|
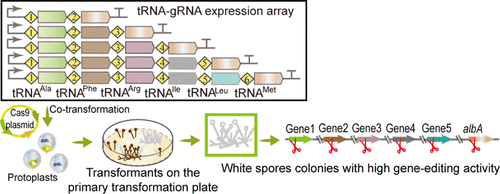
The resistance markers could ensure the entry of the CRISPR/Cas9 system into Aspergillus niger cells instead of gene editing. To increase the efficiency of positive colony screening on the primary transformation plates, we designed a visualized multigene editing system (VMS) via a unique tRNA-guide RNA (gRNA) array containing the gRNAs of a pigment gene albA and target genes. Disruption of albA produces white colonies, and the sequences of the endogenous tRNAAla, tRNAPhe, tRNAArg, tRNAIle, and tRNALeu enhance gRNA release. The disruption efficiencies of multigene were analyzed in the A. niger strain AG11 using ammA, amyA, prtT, kusA, and glaA as reporters. In white colonies on the primary transformation plates, the disruption rates of one-, two-, three-, four-, and five-target genes reached 89.2, 70.91, 50, 22.41, and 4.17%, respectively. The VMS developed here provides an effective method for screening homokaryotic multigene editing strains of A. niger.
中文翻译:

黑曲霉可视化多基因编辑系统
抗性标记可以确保 CRISPR/Cas9 系统进入黑曲霉细胞而不是基因编辑。为了提高初级转化板上阳性菌落筛选的效率,我们设计了一个可视化的多基因编辑系统 (VMS),它通过一个独特的 tRNA 引导 RNA (gRNA) 阵列包含色素基因albA和靶基因的 gRNA。albA的破坏产生白色菌落,并且内源性 tRNA Ala、tRNA Phe、tRNA Arg、tRNA Ile和 tRNA Leu的序列增强了 gRNA 的释放。在黑曲霉中分析了多基因的破坏效率使用amma、amyA、prtT、kusA和glaA作为记者菌株 AG11。在初级转化板上的白色菌落中,一、二、三、四、五靶基因的破坏率分别达到89.2、70.91、50、22.41和4.17%。这里开发的VMS为筛选黑曲霉同核多基因编辑菌株提供了一种有效的方法。
更新日期:2021-10-15
中文翻译:

黑曲霉可视化多基因编辑系统
抗性标记可以确保 CRISPR/Cas9 系统进入黑曲霉细胞而不是基因编辑。为了提高初级转化板上阳性菌落筛选的效率,我们设计了一个可视化的多基因编辑系统 (VMS),它通过一个独特的 tRNA 引导 RNA (gRNA) 阵列包含色素基因albA和靶基因的 gRNA。albA的破坏产生白色菌落,并且内源性 tRNA Ala、tRNA Phe、tRNA Arg、tRNA Ile和 tRNA Leu的序列增强了 gRNA 的释放。在黑曲霉中分析了多基因的破坏效率使用amma、amyA、prtT、kusA和glaA作为记者菌株 AG11。在初级转化板上的白色菌落中,一、二、三、四、五靶基因的破坏率分别达到89.2、70.91、50、22.41和4.17%。这里开发的VMS为筛选黑曲霉同核多基因编辑菌株提供了一种有效的方法。



























 京公网安备 11010802027423号
京公网安备 11010802027423号