Computational and Structural Biotechnology Journal ( IF 6 ) Pub Date : 2021-09-22 , DOI: 10.1016/j.csbj.2021.09.021 Shafaque Zahra 1 , Ajeet Singh 1 , Nikita Poddar 1 , Shailesh Kumar 1
|
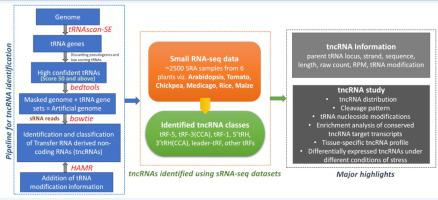
The emergence of distinct classes of non-coding RNAs has led to better insights into the eukaryotic gene regulatory networks. Amongst them, the existence of transfer RNA (tRNA)-derived non-coding RNAs (tncRNAs) demands exploration in the plant kingdom. We have designed a methodology to uncover the entire perspective of tncRNAome in plants. Using this pipeline, we have identified diverse tncRNAs with a size ranging from 14 to 50 nucleotides (nt) by utilizing 2,448 small RNA-seq samples from six angiosperms, and studied their various features, including length, codon-usage, cleavage pattern, and modified tRNA nucleosides. Codon-dependent generation of tncRNAs suggests that the tRNA cleavage is highly specific rather than random tRNA degradation. The nucleotide composition analysis of tncRNA cleavage positions indicates that they are generated through precise endoribonucleolytic cleavage machinery. Certain nucleoside modifications detected on tncRNAs were found to be conserved across the plants, and hence may influence tRNA cleavage, as well as tncRNA functions. Pathway enrichment analysis revealed that common tncRNA targets are majorly enriched during metabolic and developmental processes. Further distinct tissue-specific tncRNA clusters highlight their role in plant development. Significant number of tncRNAs differentially expressed under abiotic and biotic stresses highlights their potential role in stress resistance. In summary, this study has developed a platform that will help in the understanding of tncRNAs and their involvement in growth, development, and response to various stresses. The workflow, software package, and results are freely available at http://nipgr.ac.in/tncRNA.
中文翻译:

转移 RNA 衍生的非编码 RNA (tncRNAs):植物转录调控回路的隐藏调控
不同类别的非编码 RNA 的出现使人们对真核基因调控网络有了更好的了解。其中,转移 RNA (tRNA) 衍生的非编码 RNA (tncRNA) 的存在需要在植物界进行探索。我们设计了一种方法来揭示植物中 tncRNAome 的整个视角。使用这条管道,我们利用来自六个被子植物的 2,448 个小 RNA-seq 样本鉴定了大小范围从 14 到 50 个核苷酸 (nt) 的各种 tncRNA,并研究了它们的各种特征,包括长度、密码子使用、切割模式和修饰的 tRNA 核苷。tncRNA 的密码子依赖性生成表明 tRNA 切割是高度特异性的,而不是随机 tRNA 降解。tncRNA 切割位置的核苷酸组成分析表明它们是通过精确的内切核糖核酸切割机制产生的。发现在 tncRNA 上检测到的某些核苷修饰在植物中是保守的,因此可能会影响 tRNA 切割以及 tncRNA 功能。通路富集分析表明,常见的 tncRNA 靶点主要在代谢和发育过程中富集。进一步不同的组织特异性 tncRNA 簇突出了它们在植物发育中的作用。在非生物和生物胁迫下差异表达的大量 tncRNA 突出了它们在抗逆性中的潜在作用。总之,本研究开发了一个平台,有助于理解 tncRNA 及其参与生长、发育、以及对各种压力的反应。工作流程、软件包和结果可在以下网址免费获得http://nipgr.ac.in/tncRNA。


























 京公网安备 11010802027423号
京公网安备 11010802027423号