Journal of Molecular Graphics and Modelling ( IF 2.9 ) Pub Date : 2021-09-18 , DOI: 10.1016/j.jmgm.2021.108022 Rand Shahin 1 , Nabil N Al-Hashimi 1 , Nour El-Huda Daoud 1 , Salah Aljamal 2 , Omar Shaheen 3
|
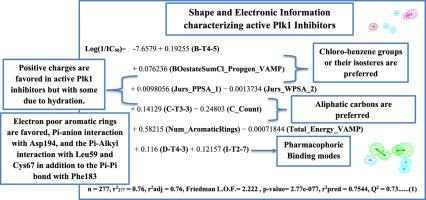
Targeting Polo-like kinase 1 (Plk1) by molecular inhibitors is being a promising approach for tumor therapy. Nevertheless, insufficient methodical analyses have been done to characterize the interactions inside the Plk1 binding pocket. In this study, an extensive combined ligand and structure-based drug design workflow was conducted to data-mine the structural requirements for Plk1 inhibition. Consequently, the binding modes of 368 previously known Plk1 inhibitors were investigated by pharmacophore generation technique. The resulted pharmacophores were engaged in the context of Genetic function algorithm (GFA) and Multiple linear regression (MLR) analyses to search for a prognostic QSAR model.
The most successful QSAR model was with statistical criteria of (r2277 = 0.76, r2adj = 0.76, r2pred = 0.75, Q2 = 0.73). Our QSAR-selected pharmacophores were validated by Receiver Operating Characteristic (ROC) curve analysis. Later on, the best QSAR model and its associated pharmacophoric hypotheses (HypoB-T4-5, HypoI-T2-7, HypoD-T4-3, and HypoC-T3-3) were used to identify new Plk1 inhibitory hits retrieved from the National Cancer Institute (NCI) database. The most potent hits exhibited experimental anti-Plk1 IC50 of 1.49, 3.79. 5.26 and 6.35 μM. Noticeably, our hits, were found to interact with the Plk1 kinase domain through some important amino acid residues namely, Cys67, Lys82, Cys133, Phe183, and Asp194.
中文翻译:

QSAR 引导的药效团模型揭示了 Polo 激酶 1 (Plk1) 抑制剂的重要结构要求
通过分子抑制剂靶向 Polo 样激酶 1 (Plk1) 是一种很有前途的肿瘤治疗方法。然而,对 Plk1 结合口袋内的相互作用进行的系统分析还不够充分。在这项研究中,进行了广泛的结合配体和基于结构的药物设计工作流程,以数据挖掘 Plk1 抑制的结构要求。因此,通过药效团生成技术研究了 368 种先前已知的 Plk1 抑制剂的结合模式。所得药效团在遗传函数算法 (GFA) 和多元线性回归 (MLR) 分析的背景下进行,以寻找预后 QSAR 模型。
最成功的 QSAR 模型的统计标准为 (r 2 277 = 0.76, r 2 adj = 0.76, r 2 pred = 0.75, Q 2 = 0.73)。我们的 QSAR 选择的药效团通过接受者操作特征 (ROC) 曲线分析进行了验证。后来,最好的 QSAR 模型及其相关的药效学假设(HypoB-T4-5、HypoI-T2-7、HypoD-T4-3 和 HypoC-T3-3)被用于识别从国家检索到的新的 Plk1 抑制命中癌症研究所 (NCI) 数据库。最有效的命中表现出 1.49、3.79 的实验性抗 Plk1 IC 50。5.26 和 6.35 微米。值得注意的是,我们的命中被发现通过一些重要的氨基酸残基与 Plk1 激酶结构域相互作用,即Cys67、Lys82、Cys133、Phe183和Asp194。


























 京公网安备 11010802027423号
京公网安备 11010802027423号