当前位置:
X-MOL 学术
›
Catal. Sci. Technol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Computational studies on the substrate specificity of an acyltransferase domain from salinomycin polyketide synthase
Catalysis Science & Technology ( IF 5 ) Pub Date : 2021-08-25 , DOI: 10.1039/d1cy00284h Huining Ji 1 , Ting Shi 1 , Lei Liu 1 , Fa Zhang 1 , Wentao Tao 1 , Qing Min 2 , Zixin Deng 1 , Linquan Bai 1 , Yilei Zhao 1 , Jianting Zheng 1, 3
Catalysis Science & Technology ( IF 5 ) Pub Date : 2021-08-25 , DOI: 10.1039/d1cy00284h Huining Ji 1 , Ting Shi 1 , Lei Liu 1 , Fa Zhang 1 , Wentao Tao 1 , Qing Min 2 , Zixin Deng 1 , Linquan Bai 1 , Yilei Zhao 1 , Jianting Zheng 1, 3
Affiliation
|
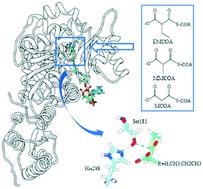
Polyketides are a large group of natural products with diverse chemical structures and biological activities. They are biosynthesized by modular polyketide synthases (PKSs) from coenzyme A (CoA) thioesters of short-chain carboxylic acids like malonyl-CoA (MCoA), methylmalonyl-CoA (MMCoA) and ethylmalonyl-CoA (EMCoA). Acyltransferase (AT) domains of modular PKSs are responsible for selecting CoA thioesters and therefore attractive targets for engineering to generate novel polyketides. Herein, molecular dynamics (MD) simulations combined with quantum mechanical/molecular-mechanical (QM/MM) calculations were conducted to dissect the substrate specificity of an AT domain from the 14th module of the salinomycin modular PKS (SalAT14), which displays a preference for its cognate substrate EMCoA over MCoA and MMCoA. Comparison of MD simulations unveiled that the hydrophobic interactions between the active site residues and the acyl groups exert a significant effect on enzyme–substrate recognition. The complex of SalAT14 and its cognate substrate EMCoA exhibited a greater tendency to stay in a conformation suitable for the reaction. QM/MM calculations demonstrated that the concerted nucleophilic attack on the thioester carbonyl group of the substrate is the rate-limiting step in the first half of transacylation. Our computational investigations revealed the structural basis of AT specificity and could potentially help the engineering of modular PKSs.
中文翻译:

盐霉素聚酮合酶酰基转移酶结构域底物特异性的计算研究
聚酮化合物是一大类具有多种化学结构和生物活性的天然产物。它们通过模块化聚酮化合物合酶 (PKS) 从短链羧酸的辅酶 A (CoA) 硫酯生物合成,如丙二酰辅酶 A (MCoA)、甲基丙二酰辅酶 A (MMCoA) 和乙基丙二酰辅酶 A (EMCoA)。模块化 PKS 的酰基转移酶 (AT) 域负责选择 CoA 硫酯,因此是用于工程生成新型聚酮化合物的有吸引力的目标。在此,分子动力学 (MD) 模拟结合量子力学/分子力学 (QM/MM) 计算,从盐霉素模块化 PKS (SalAT14) 的第 14 个模块中剖析 AT 域的底物特异性,该模块显示出偏好因为其同源底物 EMCoA 在 MCoA 和 MMCoA 之上。MD 模拟的比较表明,活性位点残基和酰基之间的疏水相互作用对酶 - 底物识别产生显着影响。SalAT14 及其同源底物 EMCoA 的复合物表现出更大的趋势,以保持适合反应的构象。QM/MM 计算表明,对底物硫酯羰基的协同亲核攻击是转酰化前半部分的限速步骤。我们的计算研究揭示了 AT 特异性的结构基础,可能有助于模块化 PKS 的工程设计。SalAT14 及其同源底物 EMCoA 的复合物表现出更大的趋势,以保持适合反应的构象。QM/MM 计算表明,对底物硫酯羰基的协同亲核攻击是转酰化前半部分的限速步骤。我们的计算研究揭示了 AT 特异性的结构基础,可能有助于模块化 PKS 的工程设计。SalAT14 及其同源底物 EMCoA 的复合物表现出更大的趋势,以保持适合反应的构象。QM/MM 计算表明,对底物硫酯羰基的协同亲核攻击是转酰化前半部分的限速步骤。我们的计算研究揭示了 AT 特异性的结构基础,可能有助于模块化 PKS 的工程设计。
更新日期:2021-09-14
中文翻译:

盐霉素聚酮合酶酰基转移酶结构域底物特异性的计算研究
聚酮化合物是一大类具有多种化学结构和生物活性的天然产物。它们通过模块化聚酮化合物合酶 (PKS) 从短链羧酸的辅酶 A (CoA) 硫酯生物合成,如丙二酰辅酶 A (MCoA)、甲基丙二酰辅酶 A (MMCoA) 和乙基丙二酰辅酶 A (EMCoA)。模块化 PKS 的酰基转移酶 (AT) 域负责选择 CoA 硫酯,因此是用于工程生成新型聚酮化合物的有吸引力的目标。在此,分子动力学 (MD) 模拟结合量子力学/分子力学 (QM/MM) 计算,从盐霉素模块化 PKS (SalAT14) 的第 14 个模块中剖析 AT 域的底物特异性,该模块显示出偏好因为其同源底物 EMCoA 在 MCoA 和 MMCoA 之上。MD 模拟的比较表明,活性位点残基和酰基之间的疏水相互作用对酶 - 底物识别产生显着影响。SalAT14 及其同源底物 EMCoA 的复合物表现出更大的趋势,以保持适合反应的构象。QM/MM 计算表明,对底物硫酯羰基的协同亲核攻击是转酰化前半部分的限速步骤。我们的计算研究揭示了 AT 特异性的结构基础,可能有助于模块化 PKS 的工程设计。SalAT14 及其同源底物 EMCoA 的复合物表现出更大的趋势,以保持适合反应的构象。QM/MM 计算表明,对底物硫酯羰基的协同亲核攻击是转酰化前半部分的限速步骤。我们的计算研究揭示了 AT 特异性的结构基础,可能有助于模块化 PKS 的工程设计。SalAT14 及其同源底物 EMCoA 的复合物表现出更大的趋势,以保持适合反应的构象。QM/MM 计算表明,对底物硫酯羰基的协同亲核攻击是转酰化前半部分的限速步骤。我们的计算研究揭示了 AT 特异性的结构基础,可能有助于模块化 PKS 的工程设计。


























 京公网安备 11010802027423号
京公网安备 11010802027423号