Molecular Cell ( IF 16.0 ) Pub Date : 2021-09-13 , DOI: 10.1016/j.molcel.2021.08.020 Andrew J Beel 1 , Maia Azubel 1 , Pierre-Jean Matteï 1 , Roger D Kornberg 1
|
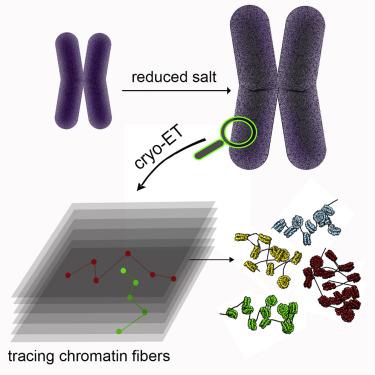
Chromatin fibers must fold or coil in the process of chromosome condensation. Patterns of coiling have been demonstrated for reconstituted chromatin, but the actual trajectories of fibers in condensed states of chromosomes could not be visualized because of the high density of the material. We have exploited partial decondensation of mitotic chromosomes to reveal their internal structure at sub-nucleosomal resolution by cryo-electron tomography, without the use of stains, fixatives, milling, or sectioning. DNA gyres around nucleosomes were visible, allowing the nucleosomes to be identified and their orientations to be determined. Linker DNA regions were traced, revealing the trajectories of the chromatin fibers. The trajectories were irregular, with almost no evidence of coiling and no short- or long-range order of the chromosomal material. The 146-bp core particle, long known as a product of nuclease digestion, is identified as the native state of the nucleosome, with no regular spacing along the chromatin fibers.
中文翻译:

有丝分裂染色体的结构
染色质纤维在染色体凝聚过程中必须折叠或卷曲。已经证明了重组染色质的卷曲模式,但由于材料的高密度,因此无法看到处于染色体凝聚状态的纤维的实际轨迹。我们利用有丝分裂染色体的部分去浓缩,通过低温电子断层扫描以亚核小体分辨率揭示其内部结构,而无需使用染色剂、固定剂、研磨或切片。核小体周围的 DNA 回旋是可见的,从而可以识别核小体并确定它们的方向。追踪接头 DNA 区域,揭示染色质纤维的轨迹。轨迹是不规则的,几乎没有盘绕的迹象,也没有染色体材料的短程或长程顺序。



























 京公网安备 11010802027423号
京公网安备 11010802027423号