当前位置:
X-MOL 学术
›
J. Chem. Theory Comput.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Biasing RNA Coarse-Grained Folding Simulations with Small-Angle X-ray Scattering Data
Journal of Chemical Theory and Computation ( IF 5.5 ) Pub Date : 2021-09-10 , DOI: 10.1021/acs.jctc.1c00441 Liuba Mazzanti 1 , Lina Alferkh 1 , Elisa Frezza 1 , Samuela Pasquali 1
Journal of Chemical Theory and Computation ( IF 5.5 ) Pub Date : 2021-09-10 , DOI: 10.1021/acs.jctc.1c00441 Liuba Mazzanti 1 , Lina Alferkh 1 , Elisa Frezza 1 , Samuela Pasquali 1
Affiliation
|
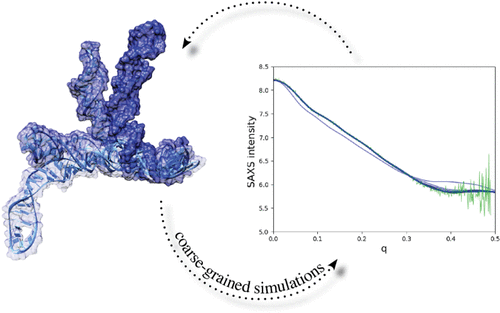
RNA molecules can easily adopt alternative structures in response to different environmental conditions. As a result, a molecule’s energy landscape is rough and can exhibit a multitude of deep basins. In the absence of a high-resolution structure, small-angle X-ray scattering data (SAXS) can narrow down the conformational space available to the molecule and be used in conjunction with physical modeling to obtain high-resolution putative structures to be further tested by experiments. Because of the low resolution of these data, it is natural to implement the integration of SAXS data into simulations using a coarse-grained representation of the molecule, allowing for much wider searches and faster evaluation of SAXS theoretical intensity curves than with atomistic models. We present here the theoretical framework and the implementation of a simulation approach based on our coarse-grained model HiRE-RNA combined with SAXS evaluations “on-the-fly” leading the simulation toward conformations agreeing with the scattering data, starting from partially folded structures as the ones that can easily be obtained from secondary structure prediction-based tools. We show on three benchmark systems how our approach can successfully achieve high-resolution structures with remarkable similarity with the native structure recovering not only the overall shape, as imposed by SAXS data, but also the details of initially missing base pairs.
中文翻译:

使用小角度 X 射线散射数据对 RNA 粗粒度折叠模拟进行偏置
RNA 分子可以很容易地采用替代结构来响应不同的环境条件。因此,一个分子的能量分布是粗糙的,可以表现出许多深盆地。在没有高分辨率结构的情况下,小角度 X 射线散射数据 (SAXS) 可以缩小分子可用的构象空间,并与物理建模结合使用以获得高分辨率推定结构以进行进一步测试通过实验。由于这些数据的低分辨率,使用分子的粗粒度表示将 SAXS 数据集成到模拟中是很自然的,与原子模型相比,允许更广泛的搜索和更快地评估 SAXS 理论强度曲线。我们在这里介绍了理论框架和模拟方法的实现,该方法基于我们的粗粒度模型 HiRE-RNA 结合 SAXS 评估“即时”引导模拟从部分折叠结构开始向与散射数据一致的构象作为可以从基于二级结构预测的工具中轻松获得的那些。我们在三个基准系统上展示了我们的方法如何成功实现与原生结构具有显着相似性的高分辨率结构,不仅恢复了 SAXS 数据强加的整体形状,还恢复了最初缺失的碱基对的细节。从部分折叠结构开始,这些结构可以很容易地从基于二级结构预测的工具中获得。我们在三个基准系统上展示了我们的方法如何成功实现与原生结构具有显着相似性的高分辨率结构,不仅恢复了 SAXS 数据强加的整体形状,还恢复了最初缺失的碱基对的细节。从部分折叠结构开始,这些结构可以很容易地从基于二级结构预测的工具中获得。我们在三个基准系统上展示了我们的方法如何成功实现与原生结构具有显着相似性的高分辨率结构,不仅恢复了 SAXS 数据强加的整体形状,还恢复了最初缺失的碱基对的细节。
更新日期:2021-10-12
中文翻译:

使用小角度 X 射线散射数据对 RNA 粗粒度折叠模拟进行偏置
RNA 分子可以很容易地采用替代结构来响应不同的环境条件。因此,一个分子的能量分布是粗糙的,可以表现出许多深盆地。在没有高分辨率结构的情况下,小角度 X 射线散射数据 (SAXS) 可以缩小分子可用的构象空间,并与物理建模结合使用以获得高分辨率推定结构以进行进一步测试通过实验。由于这些数据的低分辨率,使用分子的粗粒度表示将 SAXS 数据集成到模拟中是很自然的,与原子模型相比,允许更广泛的搜索和更快地评估 SAXS 理论强度曲线。我们在这里介绍了理论框架和模拟方法的实现,该方法基于我们的粗粒度模型 HiRE-RNA 结合 SAXS 评估“即时”引导模拟从部分折叠结构开始向与散射数据一致的构象作为可以从基于二级结构预测的工具中轻松获得的那些。我们在三个基准系统上展示了我们的方法如何成功实现与原生结构具有显着相似性的高分辨率结构,不仅恢复了 SAXS 数据强加的整体形状,还恢复了最初缺失的碱基对的细节。从部分折叠结构开始,这些结构可以很容易地从基于二级结构预测的工具中获得。我们在三个基准系统上展示了我们的方法如何成功实现与原生结构具有显着相似性的高分辨率结构,不仅恢复了 SAXS 数据强加的整体形状,还恢复了最初缺失的碱基对的细节。从部分折叠结构开始,这些结构可以很容易地从基于二级结构预测的工具中获得。我们在三个基准系统上展示了我们的方法如何成功实现与原生结构具有显着相似性的高分辨率结构,不仅恢复了 SAXS 数据强加的整体形状,还恢复了最初缺失的碱基对的细节。


























 京公网安备 11010802027423号
京公网安备 11010802027423号