Journal of Molecular Graphics and Modelling ( IF 2.9 ) Pub Date : 2021-09-05 , DOI: 10.1016/j.jmgm.2021.108011 Charmaine S M Chu 1 , Jack D Simpson 1 , Paul M O'Neill 1 , Neil G Berry 1
|
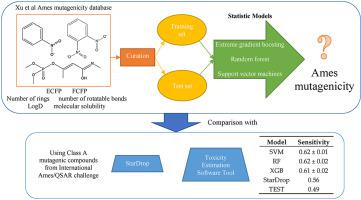
In modern drug discovery, detection of a compound's potential mutagenicity is crucial. However, the traditional method of mutagenicity detection using the Ames test is costly and time consuming as the compounds need to be synthesised and then tested and the results are not always accurate and reproducible. Therefore, it would be advantageous to develop robust in silico models which can accurately predict the mutagenicity of a compound prior to synthesis to overcome the inadequacies of the Ames test. After curation of a previously defined compound mutagenicity library, over 5000 molecules had their chemical fingerprints and molecular properties calculated. Using 8 classification modelling algorithms, including support vector machine (SVM), random forest (RF) and extreme gradient boosting (XGB), a total of 112 predictive models have been constructed. Their performance has been assessed using 10-fold cross validation and a hold-out test set and some of the top performing models have been assessed using the y-randomisation approach. As a result, we have found SVM and XGB models to have good performance during the 10-fold cross validation (AUROC >0.90, sensitivity >0.85, specificity >0.75, balanced accuracy >0.80, Kappa >0.65) and on the test set (AUROC >0.65, sensitivity >0.65, specificity >0.60, balanced accuracy >0.65, Kappa >0.30). We have also identified molecular properties that are the most influential for mutagenicity prediction when combined with chemical molecular fingerprints. Using the Class A mutagenic compounds from the Ames/QSAR International Challenge Project, we were able to verify our models perform better, predicting more mutagens correctly then the StarDrop Ames mutagenicity prediction and TEST mutagenicity prediction.
中文翻译:

机器学习——预测小分子的 Ames 致突变性
在现代药物发现中,检测化合物的潜在致突变性至关重要。然而,使用 Ames 测试的传统致突变性检测方法既昂贵又耗时,因为需要合成化合物然后进行测试,而且结果并不总是准确和可重复的。因此,开发鲁棒的计算机将是有利的可以在合成前准确预测化合物的致突变性的模型,以克服 Ames 测试的不足之处。在对先前定义的化合物致突变性文库进行管理后,计算了 5000 多个分子的化学指纹和分子特性。使用支持向量机(SVM)、随机森林(RF)和极限梯度提升(XGB)等8种分类建模算法,共构建了112个预测模型。它们的性能已使用 10 倍交叉验证和保留测试集进行了评估,并且一些表现最佳的模型已使用 y 随机化方法进行了评估。结果,我们发现 SVM 和 XGB 模型在 10 倍交叉验证期间具有良好的性能(AUROC > 0.90,敏感性 > 0.85,特异性 > 0.75,平衡准确度 >0.80,Kappa >0.65)和测试集(AUROC >0.65,敏感性 >0.65,特异性 >0.60,平衡准确度 >0.65,Kappa >0.30)。我们还确定了与化学分子指纹相结合时对诱变预测最有影响的分子特性。使用来自 Ames/QSAR 国际挑战项目的 A 类诱变化合物,我们能够验证我们的模型性能更好,比 StarDrop Ames 诱变预测和 TEST 诱变预测正确预测更多的诱变剂。我们还确定了与化学分子指纹相结合时对诱变预测最有影响的分子特性。使用来自 Ames/QSAR 国际挑战项目的 A 类诱变化合物,我们能够验证我们的模型性能更好,比 StarDrop Ames 诱变预测和 TEST 诱变预测正确预测更多的诱变剂。我们还确定了与化学分子指纹相结合时对诱变预测最有影响的分子特性。使用来自 Ames/QSAR 国际挑战项目的 A 类诱变化合物,我们能够验证我们的模型性能更好,比 StarDrop Ames 诱变预测和 TEST 诱变预测正确预测更多的诱变剂。


























 京公网安备 11010802027423号
京公网安备 11010802027423号