当前位置:
X-MOL 学术
›
Biol. Cell
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
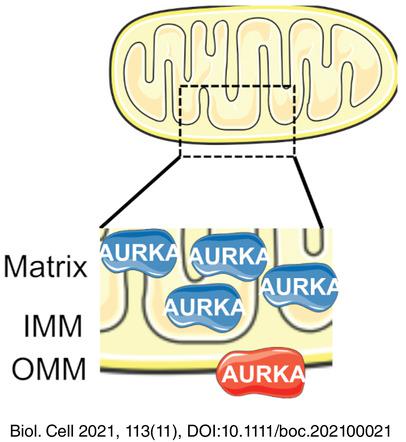
Quantitative dSTORM super-resolution microscopy localizes Aurora kinase A/AURKA in the mitochondrial matrix
Biology of the Cell ( IF 2.7 ) Pub Date : 2021-08-31 , DOI: 10.1111/boc.202100021 Béatrice Durel 1 , Charles Kervrann 2, 3 , Giulia Bertolin 4
Biology of the Cell ( IF 2.7 ) Pub Date : 2021-08-31 , DOI: 10.1111/boc.202100021 Béatrice Durel 1 , Charles Kervrann 2, 3 , Giulia Bertolin 4
Affiliation
|
Mitochondria are dynamic organelles playing essential metabolic and signaling functions in cells. Their ultrastructure has largely been investigated with electron microscopy (EM) techniques. However, quantifying protein-protein proximities using EM is extremely challenging. Super-resolution microscopy techniques as direct stochastic optical reconstruction microscopy (dSTORM) now provide a fluorescent-based, quantitative alternative to EM. Recently, super-resolution microscopy approaches including dSTORM led to valuable advances in our knowledge of mitochondrial ultrastructure, and in linking it with new insights in organelle functions. Nevertheless, dSTORM is mostly used to image integral mitochondrial proteins, and there is little or no information on proteins transiently present at this compartment. The cancer-related Aurora kinase A/AURKA is a protein localized at various subcellular locations, including mitochondria.
中文翻译:

定量 dSTORM 超分辨率显微镜在线粒体基质中定位 Aurora 激酶 A/AURKA
线粒体是在细胞中发挥重要代谢和信号功能的动态细胞器。它们的超微结构已在很大程度上用电子显微镜 (EM) 技术进行了研究。然而,使用 EM 量化蛋白质-蛋白质接近度极具挑战性。作为直接随机光学重建显微镜 (dSTORM) 的超分辨率显微镜技术现在提供了一种基于荧光的定量替代 EM。最近,包括 dSTORM 在内的超分辨率显微镜方法在我们对线粒体超微结构的认识方面取得了宝贵进展,并将其与细胞器功能的新见解联系起来。然而,dSTORM 主要用于对完整的线粒体蛋白进行成像,并且很少或没有关于瞬时存在于该隔间中的蛋白质的信息。
更新日期:2021-11-03
中文翻译:

定量 dSTORM 超分辨率显微镜在线粒体基质中定位 Aurora 激酶 A/AURKA
线粒体是在细胞中发挥重要代谢和信号功能的动态细胞器。它们的超微结构已在很大程度上用电子显微镜 (EM) 技术进行了研究。然而,使用 EM 量化蛋白质-蛋白质接近度极具挑战性。作为直接随机光学重建显微镜 (dSTORM) 的超分辨率显微镜技术现在提供了一种基于荧光的定量替代 EM。最近,包括 dSTORM 在内的超分辨率显微镜方法在我们对线粒体超微结构的认识方面取得了宝贵进展,并将其与细胞器功能的新见解联系起来。然而,dSTORM 主要用于对完整的线粒体蛋白进行成像,并且很少或没有关于瞬时存在于该隔间中的蛋白质的信息。


























 京公网安备 11010802027423号
京公网安备 11010802027423号