Journal of Molecular Biology ( IF 5.6 ) Pub Date : 2021-08-28 , DOI: 10.1016/j.jmb.2021.167219 Adi Ulman 1 , Tal Levin 2 , Bareket Dassa 1 , Aaron Javitt 1 , Assaf Kacen 1 , Merav D Shmueli 1 , Avital Eisenberg-Lerner 1 , Daoud Sheban 1 , Simon Fishllevich 3 , Emmanuel D Levy 2 , Yifat Merbl 1
|
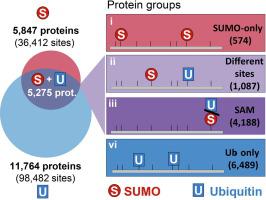
Protein modification by ubiquitin or SUMO can alter the function, stability or activity of target proteins. Previous studies have identified thousands of substrates that were modified by ubiquitin or SUMO on the same lysine residue. However, it remains unclear whether such overlap could result from a mere higher solvent accessibility, whether proteins containing those sites are associated with specific functional traits, and whether selectively perturbing their modification by ubiquitin or SUMO could result in different phenotypic outcomes. Here, we mapped reported lysine modification sites across the human proteome and found an enrichment of sites reported to be modified by both ubiquitin and SUMO. Our analysis uncovered thousands of proteins containing such sites, which we term Sites of Alternative Modification (SAMs). Among more than 36,000 sites reported to be modified by SUMO, 51.8% have also been reported to be modified by ubiquitin. SAM-containing proteins are associated with diverse biological functions including cell cycle, DNA damage, and transcriptional regulation. As such, our analysis highlights numerous proteins and pathways as putative targets for further elucidating the crosstalk between ubiquitin and SUMO. Comparing the biological and biochemical properties of SAMs versus other non-overlapping modification sites revealed that these sites were associated with altered cellular localization or abundance of their host proteins. Lastly, using S. cerevisiae as model, we show that mutating the SAM motif in a protein can influence its ubiquitination as well as its localization and abundance.
中文翻译:

从泛素和 SUMO 替代修饰位点推断的蛋白质丰度和定位改变
泛素或 SUMO 的蛋白质修饰可以改变目标蛋白质的功能、稳定性或活性。以前的研究已经确定了数千种在相同赖氨酸残基上被泛素或 SUMO 修饰的底物。然而,目前尚不清楚这种重叠是否仅由更高的溶剂可及性导致,包含这些位点的蛋白质是否与特定的功能特征相关,以及通过泛素或 SUMO 选择性干扰它们的修饰是否会导致不同的表型结果。在这里,我们绘制了人类蛋白质组中报告的赖氨酸修饰位点,并发现了据报道被泛素和 SUMO 修饰的位点的富集。我们的分析发现了数千种含有此类位点的蛋白质,我们将其称为替代修饰位点 (SAMs)。在超过 36 个中,000 个据报道被 SUMO 修饰的位点,51.8% 也被报道被泛素修饰。含 SAM 的蛋白质与多种生物学功能相关,包括细胞周期、DNA 损伤和转录调控。因此,我们的分析强调了许多蛋白质和途径作为进一步阐明泛素和 SUMO 之间串扰的假定目标。比较 SAM 的生物和生化特性与其他非重叠修饰位点相比,这些位点与细胞定位或宿主蛋白质丰度的改变有关。最后,使用酿酒酵母作为模型,我们表明突变蛋白质中的 SAM 基序可以影响其泛素化及其定位和丰度。



























 京公网安备 11010802027423号
京公网安备 11010802027423号