当前位置:
X-MOL 学术
›
ACS Chem. Biol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
A Sort-Seq Approach to the Development of Single Fluorescent Protein Biosensors
ACS Chemical Biology ( IF 4 ) Pub Date : 2021-08-25 , DOI: 10.1021/acschembio.1c00423 John N Koberstein 1 , Melissa L Stewart 1 , Taylor L Mighell 2 , Chadwick B Smith 1 , Michael S Cohen 3
ACS Chemical Biology ( IF 4 ) Pub Date : 2021-08-25 , DOI: 10.1021/acschembio.1c00423 John N Koberstein 1 , Melissa L Stewart 1 , Taylor L Mighell 2 , Chadwick B Smith 1 , Michael S Cohen 3
Affiliation
|
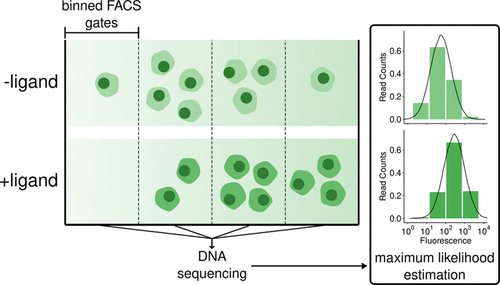
Motivated by the growing importance of single fluorescent protein biosensors (SFPBs) in biological research and the difficulty in rationally engineering these tools, we sought to increase the rate at which SFPB designs can be optimized. SFPBs generally consist of three components: a circularly permuted fluorescent protein, a ligand-binding domain, and linkers connecting the two domains. In the absence of predictive methods for biosensor engineering, most designs combining these three components will fail to produce allosteric coupling between ligand binding and fluorescence emission. While methods to construct diverse libraries with variation in the site of GFP insertion and linker sequences have been developed, the remaining bottleneck is the ability to test these libraries for functional biosensors. We address this challenge by applying a massively parallel assay termed “sort-seq,” which combines binned fluorescence-activated cell sorting, next-generation sequencing, and maximum likelihood estimation to quantify the brightness and dynamic range for many biosensor variants in parallel. We applied this method to two common biosensor optimization tasks: the choice of insertion site and optimization of linker sequences. The sort-seq assay applied to a maltose-binding protein domain-insertion library not only identified previously described high-dynamic-range variants but also discovered new functional insertion sites with diverse properties. A sort-seq assay performed on a pyruvate biosensor linker library expressed in mammalian cell culture identified linker variants with substantially improved dynamic range. Machine learning models trained on the resulting data can predict dynamic range from linker sequences. This high-throughput approach will accelerate the design and optimization of SFPBs, expanding the biosensor toolbox.
中文翻译:

用于开发单荧光蛋白生物传感器的排序序列方法
由于单一荧光蛋白生物传感器 (SFPB) 在生物研究中日益重要,以及合理设计这些工具的难度,我们寻求提高 SFPB 设计的优化速度。SFPB 通常由三个组件组成:循环排列的荧光蛋白、配体结合结构域和连接两个结构域的接头。在缺乏生物传感器工程的预测方法的情况下,结合这三个组件的大多数设计将无法在配体结合和荧光发射之间产生变构耦合。虽然已经开发了构建具有 GFP 插入位点和接头序列变化的不同文库的方法,但剩下的瓶颈是测试这些文库的功能性生物传感器的能力。我们通过应用称为“sort-seq”的大规模并行测定来应对这一挑战,该测定结合了分箱荧光激活细胞分选、下一代测序和最大似然估计,以并行量化许多生物传感器变体的亮度和动态范围。我们将该方法应用于两个常见的生物传感器优化任务:插入位点的选择和接头序列的优化。应用于麦芽糖结合蛋白结构域插入文库的 sort-seq 分析不仅鉴定了先前描述的高动态范围变体,而且还发现了具有不同特性的新功能插入位点。对在哺乳动物细胞培养物中表达的丙酮酸生物传感器接头文库进行的排序测序分析鉴定出具有显着改善的动态范围的接头变体。根据结果数据训练的机器学习模型可以预测链接序列的动态范围。这种高通量方法将加速 SFPB 的设计和优化,扩展生物传感器工具箱。
更新日期:2021-09-17
中文翻译:

用于开发单荧光蛋白生物传感器的排序序列方法
由于单一荧光蛋白生物传感器 (SFPB) 在生物研究中日益重要,以及合理设计这些工具的难度,我们寻求提高 SFPB 设计的优化速度。SFPB 通常由三个组件组成:循环排列的荧光蛋白、配体结合结构域和连接两个结构域的接头。在缺乏生物传感器工程的预测方法的情况下,结合这三个组件的大多数设计将无法在配体结合和荧光发射之间产生变构耦合。虽然已经开发了构建具有 GFP 插入位点和接头序列变化的不同文库的方法,但剩下的瓶颈是测试这些文库的功能性生物传感器的能力。我们通过应用称为“sort-seq”的大规模并行测定来应对这一挑战,该测定结合了分箱荧光激活细胞分选、下一代测序和最大似然估计,以并行量化许多生物传感器变体的亮度和动态范围。我们将该方法应用于两个常见的生物传感器优化任务:插入位点的选择和接头序列的优化。应用于麦芽糖结合蛋白结构域插入文库的 sort-seq 分析不仅鉴定了先前描述的高动态范围变体,而且还发现了具有不同特性的新功能插入位点。对在哺乳动物细胞培养物中表达的丙酮酸生物传感器接头文库进行的排序测序分析鉴定出具有显着改善的动态范围的接头变体。根据结果数据训练的机器学习模型可以预测链接序列的动态范围。这种高通量方法将加速 SFPB 的设计和优化,扩展生物传感器工具箱。


























 京公网安备 11010802027423号
京公网安备 11010802027423号