当前位置:
X-MOL 学术
›
ACS Synth. Biol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
An Integrative Toolbox for Synthetic Biology in Rhodococcus
ACS Synthetic Biology ( IF 4.7 ) Pub Date : 2021-08-24 , DOI: 10.1021/acssynbio.1c00292 James W Round 1 , Logan D Robeck 1 , Lindsay D Eltis 1
ACS Synthetic Biology ( IF 4.7 ) Pub Date : 2021-08-24 , DOI: 10.1021/acssynbio.1c00292 James W Round 1 , Logan D Robeck 1 , Lindsay D Eltis 1
Affiliation
|
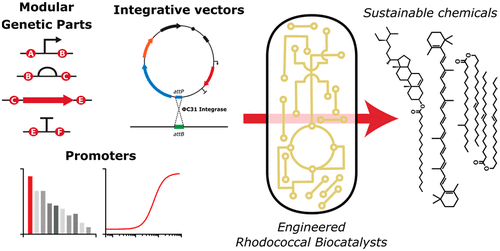
The development of microbial cell factories requires robust synthetic biology tools to reduce design uncertainty and accelerate the design-build-test-learn process. Herein, we developed a suite of integrative genetic tools to facilitate the engineering of Rhodococcus, a genus of bacteria with considerable biocatalytic potential. We first created pRIME, a modular, copy-controlled integrative-vector, to provide a robust platform for strain engineering and characterizing genetic parts. This vector was then employed to benchmark a series of strong promoters. We found PM6 to be the strongest constitutive rhodococcal promoter, 2.5- to 3-fold stronger than the next in our study, while overall promoter activities ranged 23-fold between the weakest and strongest promoters during exponential growth. Next, we used an optimized variant of PM6 to develop hybrid-promoters and integrative vectors to allow for tetracycline-inducible gene expression in Rhodococcus. The best of the resulting hybrid-promoters maintained a maximal activity of ∼50% of PM6 and displayed an induction factor of ∼40-fold. Finally, we developed and implemented a uLoop-derived Golden Gate assembly strategy for high-throughput DNA assembly in Rhodococcus. To demonstrate the utility of our approaches, pRIME was used to engineer Rhodococcus jostii RHA1 to grow on vanillin at concentrations 10-fold higher than what the wild-type strain tolerated. Overall, this study provides a suite of tools that will accelerate the engineering of Rhodococcus for various biocatalytic applications, including the sustainable production of chemicals from lignin-derived aromatics.
中文翻译:

红球菌合成生物学的综合工具箱
微生物细胞工厂的发展需要强大的合成生物学工具来减少设计不确定性并加速设计-构建-测试-学习过程。在这里,我们开发了一套综合遗传工具来促进红球菌的工程改造,红球菌是一种具有相当大的生物催化潜力的细菌。我们首先创建了 pRIME,一个模块化的、复制控制的整合载体,为应变工程和表征遗传部分提供了一个强大的平台。然后使用该载体对一系列强启动子进行基准测试。我们找到了 P M6成为最强的组成型红球菌启动子,在我们的研究中比下一个强 2.5 到 3 倍,而在指数增长期间,整体启动子活性在最弱和最强启动子之间的范围为 23 倍。接下来,我们使用 P M6的优化变体来开发杂交启动子和整合载体,以允许在红球菌中表达四环素诱导型基因。得到的最好的杂交启动子保持了约 50% 的 P M6的最大活性,并显示出约 40 倍的诱导因子。最后,我们开发并实施了一种源自 uLoop 的 Golden Gate 组装策略,用于在红球菌中进行高通量 DNA 组装。为了证明我们的方法的实用性,使用了 pRIME 来设计Rhodococcus jostii RHA1 在浓度比野生型菌株耐受浓度高 10 倍的香草醛上生长。总体而言,这项研究提供了一套工具,可以加速红球菌在各种生物催化应用中的工程设计,包括从木质素衍生的芳烃可持续生产化学品。
更新日期:2021-09-17
中文翻译:

红球菌合成生物学的综合工具箱
微生物细胞工厂的发展需要强大的合成生物学工具来减少设计不确定性并加速设计-构建-测试-学习过程。在这里,我们开发了一套综合遗传工具来促进红球菌的工程改造,红球菌是一种具有相当大的生物催化潜力的细菌。我们首先创建了 pRIME,一个模块化的、复制控制的整合载体,为应变工程和表征遗传部分提供了一个强大的平台。然后使用该载体对一系列强启动子进行基准测试。我们找到了 P M6成为最强的组成型红球菌启动子,在我们的研究中比下一个强 2.5 到 3 倍,而在指数增长期间,整体启动子活性在最弱和最强启动子之间的范围为 23 倍。接下来,我们使用 P M6的优化变体来开发杂交启动子和整合载体,以允许在红球菌中表达四环素诱导型基因。得到的最好的杂交启动子保持了约 50% 的 P M6的最大活性,并显示出约 40 倍的诱导因子。最后,我们开发并实施了一种源自 uLoop 的 Golden Gate 组装策略,用于在红球菌中进行高通量 DNA 组装。为了证明我们的方法的实用性,使用了 pRIME 来设计Rhodococcus jostii RHA1 在浓度比野生型菌株耐受浓度高 10 倍的香草醛上生长。总体而言,这项研究提供了一套工具,可以加速红球菌在各种生物催化应用中的工程设计,包括从木质素衍生的芳烃可持续生产化学品。



























 京公网安备 11010802027423号
京公网安备 11010802027423号