Computational and Structural Biotechnology Journal ( IF 6 ) Pub Date : 2021-08-19 , DOI: 10.1016/j.csbj.2021.08.029 Zhonglei Wang 1, 2 , Liyan Yang 3 , Xian-En Zhao 1
|
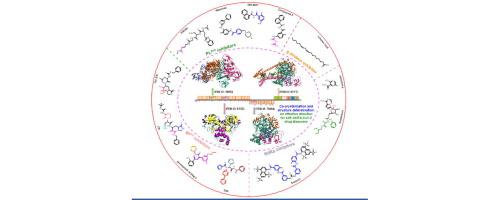
Safer and more-effective drugs are urgently needed to counter infections with the highly pathogenic SARS-CoV-2, cause of the COVID-19 pandemic. Identification of efficient inhibitors to treat and prevent SARS-CoV-2 infection is a predominant focus. Encouragingly, using X-ray crystal structures of therapeutically relevant drug targets (PLpro, Mpro, RdRp, and S glycoprotein) offers a valuable direction for anti–SARS-CoV-2 drug discovery and lead optimization through direct visualization of interactions. Computational analyses based primarily on MMPBSA calculations have also been proposed for assessing the binding stability of biomolecular structures involving the ligand and receptor. In this study, we focused on state-of-the-art X-ray co-crystal structures of the abovementioned targets complexed with newly identified small-molecule inhibitors (natural products, FDA-approved drugs, candidate drugs, and their analogues) with the assistance of computational analyses to support the precision design and screening of anti–SARS-CoV-2 drugs.
中文翻译:

共结晶和结构测定:抗SARS-CoV-2药物发现的有效方向
迫切需要更安全、更有效的药物来对抗导致 COVID-19 大流行的高致病性 SARS-CoV-2 感染。鉴定治疗和预防 SARS-CoV-2 感染的有效抑制剂是一个主要焦点。令人鼓舞的是,利用治疗相关药物靶点(PL pro、M pro 、RdRp 和 S 糖蛋白)的 X 射线晶体结构,通过直接可视化相互作用,为抗 SARS-CoV-2 药物发现和先导化合物优化提供了有价值的方向。主要基于 MMPBSA 计算的计算分析也被提议用于评估涉及配体和受体的生物分子结构的结合稳定性。在这项研究中,我们重点研究了上述靶标与新发现的小分子抑制剂(天然产物、FDA批准的药物、候选药物及其类似物)复合的最先进的X射线共晶结构,计算分析的协助支持抗 SARS-CoV-2 药物的精确设计和筛选。


























 京公网安备 11010802027423号
京公网安备 11010802027423号