当前位置:
X-MOL 学术
›
J. Comput. Chem.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Pseudopotentials for coarse-grained cross-link-assisted modeling of protein structures
Journal of Computational Chemistry ( IF 3 ) Pub Date : 2021-08-17 , DOI: 10.1002/jcc.26736 Mateusz Kogut 1 , Zhou Gong 2 , Chun Tang 3 , Adam Liwo 1
Journal of Computational Chemistry ( IF 3 ) Pub Date : 2021-08-17 , DOI: 10.1002/jcc.26736 Mateusz Kogut 1 , Zhou Gong 2 , Chun Tang 3 , Adam Liwo 1
Affiliation
|
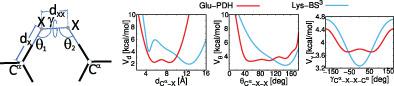
Pseudopotentials for the chemical cross-links comprising the glutamic- and aspartic-acid side chains bridged with adipic- (ADH) or pimelic-acid hydrazide (PDH), and the lysine side chains bridged with glutaric (BS2G) or suberic acid (BS3) for coarse-grained cross-link-assisted simulations were determined by canonical molecular dynamics with the Amber14sb force field. The potentials depend on the distance between side-chain ends and on side-chain orientation, this preventing from making cross-link contacts across the globule in simulations. The potentials were implemented in the UNRES coarse-grained force field and their effect on the quality of models was assessed with 11 monomeric and 1 dimeric proteins, using synthetic or experimental cross-link data. Simulations with the new potentials resulted in improvement of the generated models compared to unrestrained simulations in more instances compared to those with the statistical potentials.
中文翻译:

蛋白质结构粗粒度交联辅助建模的赝势
化学交联的赝势包括与己二酸 (ADH) 或庚二酸酰肼 (PDH) 桥接的谷氨酸和天冬氨酸侧链,以及与戊二酸 (BS 2 G) 或辛二酸桥接的赖氨酸侧链 ( BS 3) 对于粗粒度的交联辅助模拟,由典型分子动力学和 Amber14sb 力场确定。电位取决于侧链末端之间的距离和侧链方向,这防止了在模拟中跨球体进行交联接触。在 UNRES 粗粒度力场中实施了电位,并使用合成或实验交联数据,用 11 种单体和 1 种二聚体蛋白评估了它们对模型质量的影响。与具有统计潜力的模拟相比,在更多实例中,具有新潜力的模拟导致生成的模型得到改进。
更新日期:2021-09-24
中文翻译:

蛋白质结构粗粒度交联辅助建模的赝势
化学交联的赝势包括与己二酸 (ADH) 或庚二酸酰肼 (PDH) 桥接的谷氨酸和天冬氨酸侧链,以及与戊二酸 (BS 2 G) 或辛二酸桥接的赖氨酸侧链 ( BS 3) 对于粗粒度的交联辅助模拟,由典型分子动力学和 Amber14sb 力场确定。电位取决于侧链末端之间的距离和侧链方向,这防止了在模拟中跨球体进行交联接触。在 UNRES 粗粒度力场中实施了电位,并使用合成或实验交联数据,用 11 种单体和 1 种二聚体蛋白评估了它们对模型质量的影响。与具有统计潜力的模拟相比,在更多实例中,具有新潜力的模拟导致生成的模型得到改进。


























 京公网安备 11010802027423号
京公网安备 11010802027423号