当前位置:
X-MOL 学术
›
ACS Synth. Biol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Advances in Genome-Scale Metabolic Modeling toward Microbial Community Analysis of the Human Microbiome
ACS Synthetic Biology ( IF 4.7 ) Pub Date : 2021-08-17 , DOI: 10.1021/acssynbio.1c00140 Elif Esvap 1 , Kutlu O Ulgen 1
ACS Synthetic Biology ( IF 4.7 ) Pub Date : 2021-08-17 , DOI: 10.1021/acssynbio.1c00140 Elif Esvap 1 , Kutlu O Ulgen 1
Affiliation
|
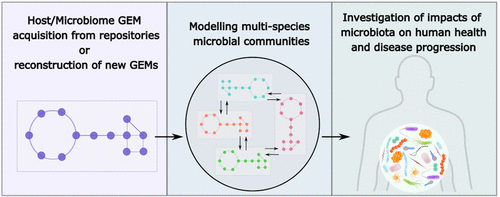
A genome-scale metabolic model (GEM) represents metabolic pathways of an organism in a mathematical form and can be built using biochemistry and genome annotation data. GEMs are invaluable for understanding organisms since they analyze the metabolic capabilities and behaviors quantitatively and can predict phenotypes. The development of high-throughput data collection techniques led to an immense increase in omics data such as metagenomics, which expand our knowledge on the human microbiome, but this also created a need for systematic analysis of these data. In recent years, GEMs have also been reconstructed for microbial species, including human gut microbiota, and methods for the analysis of microbial communities have been developed to examine the interaction between the organisms or the host. The purpose of this review is to provide a comprehensive guide for the applications of GEMs in microbial community analysis. Starting with GEM repositories, automatic GEM reconstruction tools, and quality control of models, this review will give insights into microbe–microbe and microbe–host interaction predictions and optimization of microbial community models. Recent studies that utilize microbial GEMs and personalized models to infer the influence of microbiota on human diseases such as inflammatory bowel diseases (IBD) or Parkinson’s disease are exemplified. Being powerful system biology tools for both species-level and community-level analysis of microbes, GEMs integrated with omics data and machine learning techniques will be indispensable for studying the microbiome and their effects on human physiology as well as for deciphering the mechanisms behind human diseases.
中文翻译:

人类微生物组微生物群落分析的基因组规模代谢建模进展
基因组规模的代谢模型 (GEM) 以数学形式表示生物体的代谢途径,并且可以使用生物化学和基因组注释数据构建。GEM 对于理解生物体非常宝贵,因为它们定量分析代谢能力和行为并可以预测表型。高通量数据收集技术的发展导致宏基因组学等组学数据的大量增加,这扩展了我们对人类微生物组的了解,但这也产生了对这些数据进行系统分析的需求。近年来,还针对微生物物种(包括人类肠道微生物群)重建了 GEM,并且已经开发出分析微生物群落的方法来检查生物体或宿主之间的相互作用。本综述的目的是为 GEM 在微生物群落分析中的应用提供全面的指导。从 GEM 存储库、自动 GEM 重建工具和模型质量控制开始,本综述将深入了解微生物-微生物和微生物-宿主相互作用的预测和微生物群落模型的优化。最近的研究利用微生物 GEM 和个性化模型来推断微生物群对炎症性肠病 (IBD) 或帕金森病等人类疾病的影响。作为强大的系统生物学工具,用于微生物的物种水平和群落水平分析,
更新日期:2021-09-17
中文翻译:

人类微生物组微生物群落分析的基因组规模代谢建模进展
基因组规模的代谢模型 (GEM) 以数学形式表示生物体的代谢途径,并且可以使用生物化学和基因组注释数据构建。GEM 对于理解生物体非常宝贵,因为它们定量分析代谢能力和行为并可以预测表型。高通量数据收集技术的发展导致宏基因组学等组学数据的大量增加,这扩展了我们对人类微生物组的了解,但这也产生了对这些数据进行系统分析的需求。近年来,还针对微生物物种(包括人类肠道微生物群)重建了 GEM,并且已经开发出分析微生物群落的方法来检查生物体或宿主之间的相互作用。本综述的目的是为 GEM 在微生物群落分析中的应用提供全面的指导。从 GEM 存储库、自动 GEM 重建工具和模型质量控制开始,本综述将深入了解微生物-微生物和微生物-宿主相互作用的预测和微生物群落模型的优化。最近的研究利用微生物 GEM 和个性化模型来推断微生物群对炎症性肠病 (IBD) 或帕金森病等人类疾病的影响。作为强大的系统生物学工具,用于微生物的物种水平和群落水平分析,


























 京公网安备 11010802027423号
京公网安备 11010802027423号