Journal of Molecular Graphics and Modelling ( IF 2.9 ) Pub Date : 2021-08-14 , DOI: 10.1016/j.jmgm.2021.108009 Ayisha Zia 1 , Sajid Rashid 1
|
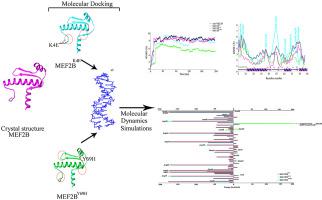
Transcriptional coactivator myocyte enhancer factor 2B (MEF2B) mutations are the most common cause of germinal center-derived B-cell non-Hodgkin lymphoma. Despite well-established contributions in lymphomagenesis, the structure-function paradigms of these mutations are largely unknown. Here through in silico approaches, we present structural evaluation of two reported missense variants (K4E and Y69H) in MEF2B to investigate their impact on DNA-binding through molecular dynamics simulation assays. Notably, MEF2B-specific MADs box domain (Lys23, Arg24 and Lys31) and N-terminal loop residues (Gly2, Arg3, Lys4, Lys5, Ile6 and Asn13) contribute in DNA binding, while in MEF2BK4E, DNA binding is facilitated by Gly2, Arg3 and Arg91 (α3) residues. Conversely, in MEF2BY69H, Arg3, Lys5, Ser78, Arg79 and Asn81 residues mediate DNA binding. DNA binding induces pronounced conformational readjustments in MEF2BWT-specific α1-N-terminal loop region, while MEF2BY69H and MEF2BK4E exhibit fluctuations in both α1 and α3. Hydrogen (H)-bond occupancy analysis reveals a similar DNA binding behavior for MEF2WT and MEF2BY69H, compared to MEF2BK4E structure. The Anisotropic Network Model analysis depicts α1 and α3 as more fluctuant regions in MEF2BK4E as compared to other systems. MEF2BWT and MEF2BK4E, Tyr69 residue is involved in p300 binding thus possible influence of Y69H variation in the functions other than DNA binding, such as p300 co-activator recruitment may explain the reduced transcriptional activation of MEF2BY69H. Thus, present study may provide a structural basis of DNA recognition by pinpointing the underlying conformational changes in the dynamics of MEF2BK4E, MEF2BY69H, and MEF2BWT structures that may contribute in the identification of novel therapeutic strategies for lymphomagenesis.
中文翻译:

由于 Y69H 和 K4E 变体,MEF2B-DNA 结合界面中的系统转换建模分析
转录共激活肌细胞增强因子 2B (MEF2B) 突变是生发中心衍生的 B 细胞非霍奇金淋巴瘤的最常见原因。尽管在淋巴瘤发生中有公认的贡献,但这些突变的结构 - 功能范例在很大程度上是未知的。在这里,通过计算机方法,我们对 MEF2B 中两个报告的错义变体(K4E 和 Y69H)进行了结构评估,以通过分子动力学模拟分析研究它们对 DNA 结合的影响。值得注意的是,MEF2B 特异性 MADs 盒结构域(Lys23、Arg24 和 Lys31)和N端环残基(Gly2、Arg3、Lys4、Lys5、Ile6 和 Asn13)有助于 DNA 结合,而在 MEF2B K4E中,DNA 结合由 Gly2 促进, Arg3 和 Arg91 (α3) 残基。相反,在 MEF2B Y69H、Arg3、Lys5、Ser78、Arg79 和 Asn81 残基介导 DNA 结合。DNA 结合在 MEF2B WT特异性 α1- N末端环区诱导显着的构象重新调整,而 MEF2B Y69H和 MEF2B K4E在 α1 和 α3 中都表现出波动。与 MEF2B K4E结构相比,氢 (H) 键占用分析揭示了 MEF2 WT和 MEF2B Y69H的相似 DNA 结合行为。与其他系统相比,各向异性网络模型分析将 α1 和 α3 描述为 MEF2B K4E中波动更大的区域。MEF2B WT和 MEF2B K4E, Tyr69 残基参与 p300 结合,因此 Y69H 变异可能影响除 DNA 结合以外的其他功能,例如 p300 共激活因子募集可以解释 MEF2B Y69H的转录激活减少。因此,本研究可以通过查明 MEF2B K4E、MEF2B Y69H和 MEF2B WT结构的动力学中潜在的构象变化来提供 DNA 识别的结构基础,这可能有助于确定淋巴瘤发生的新治疗策略。



























 京公网安备 11010802027423号
京公网安备 11010802027423号