当前位置:
X-MOL 学术
›
ACS Synth. Biol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
History-Driven Genetic Modification Design Technique Using a Domain-Specific Lexical Model for the Acceleration of DBTL Cycles for Microbial Cell Factories
ACS Synthetic Biology ( IF 4.7 ) Pub Date : 2021-08-05 , DOI: 10.1021/acssynbio.1c00234 Shiori Nakazawa 1 , Osamu Imaichi 1 , Takahisa Kogure 2 , Takeshi Kubota 2 , Koichi Toyoda 2 , Masako Suda 2 , Masayuki Inui 2 , Kiyoto Ito 1 , Tomokazu Shirai 3 , Michihiro Araki 4, 5, 6
ACS Synthetic Biology ( IF 4.7 ) Pub Date : 2021-08-05 , DOI: 10.1021/acssynbio.1c00234 Shiori Nakazawa 1 , Osamu Imaichi 1 , Takahisa Kogure 2 , Takeshi Kubota 2 , Koichi Toyoda 2 , Masako Suda 2 , Masayuki Inui 2 , Kiyoto Ito 1 , Tomokazu Shirai 3 , Michihiro Araki 4, 5, 6
Affiliation
|
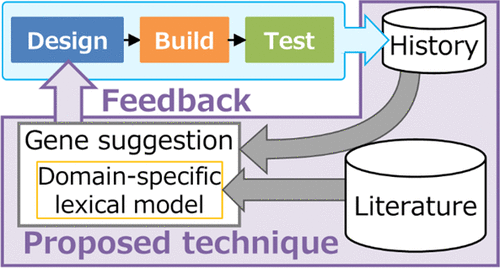
The development of microbes for conducting bioprocessing via synthetic biology involves design–build–test–learn (DBTL) cycles. To aid the designing step, we developed a computational technique that suggests next genetic modifications on the basis of relatedness to the user’s design history of genetic modifications accumulated through former DBTL cycles conducted by the user. This technique, which comprehensively retrieves well-known designs related to the history, involves searching text for previous literature and then mining genes that frequently co-occur in the literature with those modified genes. We further developed a domain-specific lexical model that weights literature that is more related to the domain of metabolic engineering to emphasize genes modified for bioprocessing. Our technique made a suggestion by using a history of creating a Corynebacterium glutamicum strain producing shikimic acid that had 18 genetic modifications. Inspired by the suggestion, eight genes were considered by biologists for further modification, and modifying four of these genes proved experimentally efficient in increasing the production of shikimic acid. These results indicated that our proposed technique successfully utilized the former cycles to suggest relevant designs that biologists considered worth testing. Comprehensive retrieval of well-tested designs will help less-experienced researchers overcome the entry barrier as well as inspire experienced researchers to formulate design concepts that have been overlooked or suspended. This technique will aid DBTL cycles by feeding histories back to the next genetic design, thereby complementing the designing step.
中文翻译:

历史驱动的基因修饰设计技术,使用特定领域的词汇模型来加速微生物细胞工厂的 DBTL 循环
用于进行生物加工的微生物的开发合成生物学涉及设计-构建-测试-学习(DBTL)循环。为了帮助设计步骤,我们开发了一种计算技术,该技术根据与用户在以前的 DBTL 循环中积累的遗传修改的设计历史的相关性来建议下一个遗传修改。该技术全面检索与历史相关的知名设计,包括搜索先前文献的文本,然后挖掘文献中经常与这些修饰基因共存的基因。我们进一步开发了一个特定领域的词汇模型,该模型对与代谢工程领域更相关的文献进行加权,以强调为生物加工而修改的基因。我们的技术利用创造谷氨酸棒杆菌的历史提出了一个建议产生莽草酸的菌株具有 18 种基因修饰。受该建议的启发,生物学家考虑对八个基因进行进一步修饰,其中四个基因的修饰在实验上证明在增加莽草酸产量方面是有效的。这些结果表明,我们提出的技术成功地利用了以前的周期来提出生物学家认为值得测试的相关设计。全面检索经过良好测试的设计将帮助经验不足的研究人员克服进入障碍,并激发有经验的研究人员制定被忽视或暂停的设计概念。这种技术将通过将历史反馈给下一个基因设计来帮助 DBTL 循环,从而补充设计步骤。
更新日期:2021-09-17
中文翻译:

历史驱动的基因修饰设计技术,使用特定领域的词汇模型来加速微生物细胞工厂的 DBTL 循环
用于进行生物加工的微生物的开发合成生物学涉及设计-构建-测试-学习(DBTL)循环。为了帮助设计步骤,我们开发了一种计算技术,该技术根据与用户在以前的 DBTL 循环中积累的遗传修改的设计历史的相关性来建议下一个遗传修改。该技术全面检索与历史相关的知名设计,包括搜索先前文献的文本,然后挖掘文献中经常与这些修饰基因共存的基因。我们进一步开发了一个特定领域的词汇模型,该模型对与代谢工程领域更相关的文献进行加权,以强调为生物加工而修改的基因。我们的技术利用创造谷氨酸棒杆菌的历史提出了一个建议产生莽草酸的菌株具有 18 种基因修饰。受该建议的启发,生物学家考虑对八个基因进行进一步修饰,其中四个基因的修饰在实验上证明在增加莽草酸产量方面是有效的。这些结果表明,我们提出的技术成功地利用了以前的周期来提出生物学家认为值得测试的相关设计。全面检索经过良好测试的设计将帮助经验不足的研究人员克服进入障碍,并激发有经验的研究人员制定被忽视或暂停的设计概念。这种技术将通过将历史反馈给下一个基因设计来帮助 DBTL 循环,从而补充设计步骤。


























 京公网安备 11010802027423号
京公网安备 11010802027423号