Computers in Biology and Medicine ( IF 7.7 ) Pub Date : 2021-07-21 , DOI: 10.1016/j.compbiomed.2021.104650 Om Prakash Singh 1 , Marta Vallejo 2 , Ismail M El-Badawy 3 , Ali Aysha 1 , Jagannathan Madhanagopal 4 , Ahmad Athif Mohd Faudzi 5
|
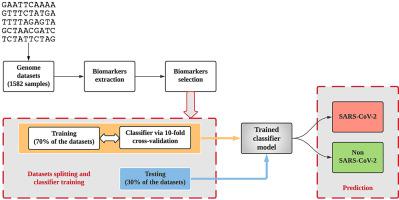
Due to the continued evolution of the SARS-CoV-2 pandemic, researchers worldwide are working to mitigate, suppress its spread, and better understand it by deploying digital signal processing (DSP) and machine learning approaches. This study presents an alignment-free approach to classify the SARS-CoV-2 using complementary DNA, which is DNA synthesized from the single-stranded RNA virus. Herein, a total of 1582 samples, with different lengths of genome sequences from different regions, were collected from various data sources and divided into a SARS-CoV-2 and a non-SARS-CoV-2 group. We extracted eight biomarkers based on three-base periodicity, using DSP techniques, and ranked those based on a filter-based feature selection. The ranked biomarkers were fed into k-nearest neighbor, support vector machines, decision trees, and random forest classifiers for the classification of SARS-CoV-2 from other coronaviruses. The training dataset was used to test the performance of the classifiers based on accuracy and F-measure via 10-fold cross-validation. Kappa-scores were estimated to check the influence of unbalanced data. Further, 10 × 10 cross-validation paired t-test was utilized to test the best model with unseen data. Random forest was elected as the best model, differentiating the SARS-CoV-2 coronavirus from other coronaviruses and a control a group with an accuracy of 97.4 %, sensitivity of 96.2 %, and specificity of 98.2 %, when tested with unseen samples. Moreover, the proposed algorithm was computationally efficient, taking only 0.31 s to compute the genome biomarkers, outperforming previous studies.
中文翻译:

使用机器学习算法对 SARS-CoV-2 和非 SARS-CoV-2 进行分类
由于SARS-CoV-2大流行的持续演变,全球研究人员正在努力通过部署数字信号处理 ( DSP ) 和机器学习方法来减轻、抑制其传播并更好地了解它。本研究提出了一种使用互补 DNA 对SARS-CoV-2进行分类的无比对方法,互补DNA是由单链RNA病毒合成的DNA 。在此,从各种数据源中收集了来自不同地区的具有不同长度基因组序列的总共 1582 个样本,并将其分为SARS-CoV-2和非 SARS-CoV-2团体。我们使用DSP技术基于三碱基周期性提取了八个生物标志物,并根据基于过滤器的特征选择对它们进行排名。排序后的生物标志物被输入到 k 最近邻、支持向量机、决策树和随机森林分类器中,用于将SARS-CoV-2与其他冠状病毒分类。训练数据集用于通过 10 折交叉验证基于准确性和 F 度量来测试分类器的性能。估计 Kappa 分数以检查不平衡数据的影响。此外,使用 10 × 10 交叉验证配对t检验来测试具有未见数据的最佳模型。随机森林被选为最佳模型,区分了SARS-CoV-2当用看不见的样本进行测试时,来自其他冠状病毒和对照组的冠状病毒的准确度为 97.4%,灵敏度为 96.2%,特异性为 98.2%。此外,所提出的算法计算效率高,仅需 0.31 秒即可计算出基因组生物标志物,优于之前的研究。


























 京公网安备 11010802027423号
京公网安备 11010802027423号