Current Biology ( IF 9.2 ) Pub Date : 2021-07-21 , DOI: 10.1016/j.cub.2021.06.056 Hiroshi Kiyonari 1 , Mari Kaneko 1 , Takaya Abe 1 , Aki Shiraishi 1 , Riko Yoshimi 1 , Ken-Ichi Inoue 1 , Yasuhide Furuta 2
|
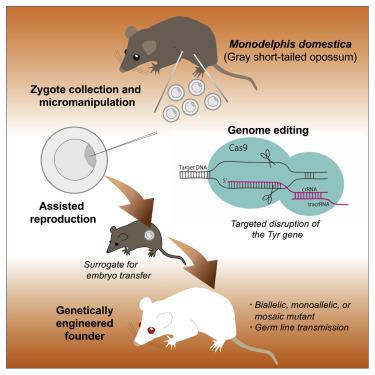
Marsupials represent one of three extant mammalian subclasses with very unique characteristics not shared by other mammals. Most notably, much of the development of neonates immaturely born after a relatively short gestation takes place in the external environment. Among marsupials, the gray short-tailed opossum (Monodelphis domestica; hereafter “the opossum”) is one of very few established laboratory models. Due to many biologically unique characteristics and experimentally advantageous features, the opossum is used as a prototype species for basic research on marsupial biology.1,2 However, in vivo studies of gene function in the opossum, and thus marsupials in general, lag far behind those of eutherian mammals due to the lack of reliable means to manipulate their genomes. In this study, we describe the successful generation of genome edited opossums by a combination of refined methodologies in reproductive biology and embryo manipulation. We took advantage of the opossum’s resemblance to popular rodent models, such as the mouse and rat, in body size and breeding characteristics. First, we established a tractable pipeline of reproductive technologies, from induction of ovulation, timed copulation, and zygote collection to embryo transfer to pseudopregnant females, that warrant an essential platform to manipulate opossum zygotes. Further, we successfully demonstrated the generation of gene knockout opossums at the Tyr locus by microinjection of pronuclear stage zygotes using CRISPR/Cas9 genome editing, along with germline transmission of the edited alleles to the F1 generation. This study provides a critical foundation for venues to expand mammalian reverse genetics into the metatherian subclass.
中文翻译:

CRISPR/Cas9 基因组编辑对有袋动物 Monodelphis domestica 的靶向基因破坏
有袋动物代表了三个现存哺乳动物亚类之一,具有其他哺乳动物所不具备的非常独特的特征。最值得注意的是,在相对较短的妊娠期后出生的未成熟新生儿的大部分发育发生在外部环境中。在有袋动物中,灰色短尾负鼠(Monodelphis domestica;以下简称“负鼠”)是极少数已建立的实验室模型之一。由于许多生物学上独特的特性和实验上的优势,负鼠被用作有袋类生物学基础研究的原型物种。1 , 2然而,在体内由于缺乏可靠的方法来操纵它们的基因组,对负鼠以及一般有袋动物基因功能的研究远远落后于真兽类哺乳动物的研究。在这项研究中,我们描述了通过结合生殖生物学和胚胎操作中的改进方法成功生成基因组编辑的负鼠。我们利用负鼠在体型和繁殖特征方面与流行的啮齿动物模型(例如小鼠和大鼠)的相似之处。首先,我们建立了一条易于处理的生殖技术管道,从诱导排卵、定时交配和受精卵收集到胚胎移植到假孕雌性,这保证了操纵负鼠受精卵的重要平台。此外,我们成功地证明了基因敲除负鼠的产生Tyr基因座通过使用 CRISPR/Cas9 基因组编辑显微注射原核期受精卵,以及将编辑的等位基因种系传递到 F1 代。这项研究为将哺乳动物反向遗传学扩展到元兽亚类的场所提供了重要基础。


























 京公网安备 11010802027423号
京公网安备 11010802027423号