当前位置:
X-MOL 学术
›
ACS Synth. Biol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Reprogramming Gene Expression by Targeting RNA-Based Interactions: A Novel Pipeline Utilizing RNA Array Technology
ACS Synthetic Biology ( IF 4.7 ) Pub Date : 2021-07-20 , DOI: 10.1021/acssynbio.0c00603 Charlotte A Henderson 1 , Helen A Vincent 1 , Anastasia J Callaghan 1
ACS Synthetic Biology ( IF 4.7 ) Pub Date : 2021-07-20 , DOI: 10.1021/acssynbio.0c00603 Charlotte A Henderson 1 , Helen A Vincent 1 , Anastasia J Callaghan 1
Affiliation
|
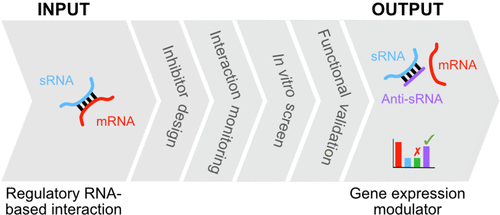
Regulatory RNA-based interactions are critical for coordinating gene expression and are increasingly being targeted in synthetic biology, antimicrobial, and therapeutic fields. Bacterial trans-encoded small RNAs (sRNAs) regulate the translation and/or stability of mRNA targets through base-pairing interactions. These interactions are often integral to complex gene circuits which coordinate critical bacterial processes. The ability to predictably modulate these gene circuits has potential for reprogramming gene expression for synthetic biology and antibacterial purposes. Here, we present a novel pipeline for targeting such RNA-based interactions with antisense oligonucleotides (ASOs) in order to reprogram gene expression. As proof-of-concept, we selected sRNA–mRNA interactions that are central to the Vibrio cholerae quorum sensing pathway, required for V. cholerae pathogenesis, as a regulatory RNA-based interaction input. We rationally designed anti-sRNA ASOs to target the sRNAs and synthesized them as peptide nucleic acids (PNAs). Next, we devised an RNA array-based interaction assay to allow screening of the anti-sRNA ASOs in vitro. Finally, an Escherichia coli-based gene expression reporter assay was developed and used to validate anti-sRNA ASO regulatory activity in a cellular environment. The output from the pipeline was an anti-sRNA ASO that targets sRNAs to inhibit sRNA–mRNA interactions and modulate gene expression. This anti-sRNA ASO has potential for reprogramming gene expression for synthetic biology and/or antibacterial purposes. We anticipate that this pipeline will find widespread use in fields targeting RNA-based interactions as modulators of gene expression.
中文翻译:

通过靶向基于 RNA 的相互作用重编程基因表达:利用 RNA 阵列技术的新型管道
基于调节性 RNA 的相互作用对于协调基因表达至关重要,并且越来越多地成为合成生物学、抗菌和治疗领域的目标。细菌反式编码小 RNA (sRNA) 通过碱基配对相互作用调节 mRNA 靶标的翻译和/或稳定性。这些相互作用通常是协调关键细菌过程的复杂基因回路的组成部分。可预测地调节这些基因回路的能力有可能为合成生物学和抗菌目的重新编程基因表达。在这里,我们提出了一种新的管道,用于针对这种基于 RNA 的与反义寡核苷酸 (ASO) 的相互作用,以重新编程基因表达。作为概念验证,我们选择了对霍乱弧菌发病机制所需的霍乱弧菌群体感应途径,作为基于 RNA 的调节相互作用输入。我们合理地设计了抗 sRNA ASO 以靶向 sRNA 并将它们合成为肽核酸 (PNA)。接下来,我们设计了一种基于 RNA 阵列的相互作用测定,以允许在体外筛选抗 sRNA ASO 。最后,大肠杆菌基于基因表达报告基因的分析被开发并用于验证细胞环境中的抗 sRNA ASO 调节活性。该管道的输出是一种抗 sRNA ASO,它以 sRNA 为目标,抑制 sRNA-mRNA 相互作用并调节基因表达。这种抗 sRNA ASO 具有为合成生物学和/或抗菌目的重新编程基因表达的潜力。我们预计,该管道将在靶向基于 RNA 的相互作用作为基因表达调节剂的领域得到广泛应用。
更新日期:2021-08-20
中文翻译:

通过靶向基于 RNA 的相互作用重编程基因表达:利用 RNA 阵列技术的新型管道
基于调节性 RNA 的相互作用对于协调基因表达至关重要,并且越来越多地成为合成生物学、抗菌和治疗领域的目标。细菌反式编码小 RNA (sRNA) 通过碱基配对相互作用调节 mRNA 靶标的翻译和/或稳定性。这些相互作用通常是协调关键细菌过程的复杂基因回路的组成部分。可预测地调节这些基因回路的能力有可能为合成生物学和抗菌目的重新编程基因表达。在这里,我们提出了一种新的管道,用于针对这种基于 RNA 的与反义寡核苷酸 (ASO) 的相互作用,以重新编程基因表达。作为概念验证,我们选择了对霍乱弧菌发病机制所需的霍乱弧菌群体感应途径,作为基于 RNA 的调节相互作用输入。我们合理地设计了抗 sRNA ASO 以靶向 sRNA 并将它们合成为肽核酸 (PNA)。接下来,我们设计了一种基于 RNA 阵列的相互作用测定,以允许在体外筛选抗 sRNA ASO 。最后,大肠杆菌基于基因表达报告基因的分析被开发并用于验证细胞环境中的抗 sRNA ASO 调节活性。该管道的输出是一种抗 sRNA ASO,它以 sRNA 为目标,抑制 sRNA-mRNA 相互作用并调节基因表达。这种抗 sRNA ASO 具有为合成生物学和/或抗菌目的重新编程基因表达的潜力。我们预计,该管道将在靶向基于 RNA 的相互作用作为基因表达调节剂的领域得到广泛应用。


























 京公网安备 11010802027423号
京公网安备 11010802027423号