当前位置:
X-MOL 学术
›
FEBS Open Bio
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Cloning, expression, and in silico structural modeling of cholesterol oxidase of Acinetobacter sp. strain RAMD in E. coli
FEBS Open Bio ( IF 2.6 ) Pub Date : 2021-07-17 , DOI: 10.1002/2211-5463.13254 Hoda E Mahmoud 1 , Shaymaa W El-Far 2 , Amira M Embaby 1
FEBS Open Bio ( IF 2.6 ) Pub Date : 2021-07-17 , DOI: 10.1002/2211-5463.13254 Hoda E Mahmoud 1 , Shaymaa W El-Far 2 , Amira M Embaby 1
Affiliation
|
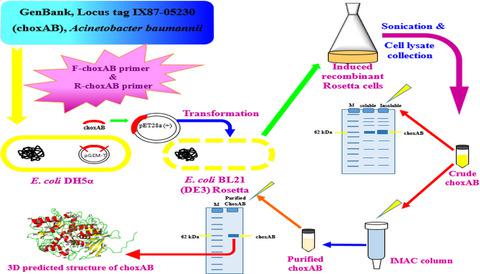
Cholesterol oxidases (CHOXs) are flavin-adenine dinucleotide-dependent oxidoreductases with a range of biotechnological applications. There remains an urgent need to identify novel CHOX family members to meet the demands of enzyme markets worldwide. Here, we report the cloning, heterologous expression, and structural modeling of the cholesterol oxidase of Acinetobacter sp. strain RAMD. The cholesterol oxidase gene was cloned and expressed in pGEM®-T and pET-28a(+) vectors, respectively, using a gene-specific primer based on the putative cholesterol oxidase ORF of Acinetobacter baumannii strain AB030 (GenBank [gb] locus tag: IX87_05230). The obtained nucleotide sequence (1671 bp, gb: MK575469.2), translated to a protein designated choxAB (556 amino acids), was overexpressed as inclusion bodies (IBs) (MW ˜ 62 kDa) in 1 mm IPTG-induced Escherichia coli BL21 (DE3) Rosetta cells. The optimized expression conditions (1 mm IPTG with 2% [v/v] glycerol and at room temperature) yielded soluble active choxAB of 0.45 U·mL−1, with 56.25-fold enhancement. The recombinant choxAB was purified to homogeneity using Ni2+-affinity agarose column with specific activity (0.054 U·mg−1), yield (8.1%), and fold purification (11.69). Capillary isoelectric-focusing indicated pI of 8.77 for choxAB. LC-MS/MS confirmed the IBs (62 kDa), with 82.6% of the covered sequence being exclusive to A. baumannii cholesterol oxidase (UniProtKB: A0A0E1FG24). The 3D structure of choxAB was predicted using the LOMETS webtool with the cholesterol oxidase template of Streptomyces sp. SA-COO (PDB: 2GEW). The predicted secondary structure included 18 α-helices and 12 β-strands, a predicted catalytic triad (E220, H380, and N514), and a conserved FAD-binding sequence (GSGFGGSVSACRLTEKG). Future studies should consider fusion to solubilization tags and switching to the expression host Pichia pastoris to reduce IB formation.
中文翻译:

不动杆菌属的胆固醇氧化酶的克隆、表达和计算机结构建模。大肠杆菌中的 RAMD 菌株
胆固醇氧化酶 (CHOX) 是黄素腺嘌呤二核苷酸依赖性氧化还原酶,具有一系列生物技术应用。仍然迫切需要确定新的 CHOX 家族成员以满足全球酶市场的需求。在这里,我们报告了不动杆菌属的胆固醇氧化酶的克隆、异源表达和结构建模。应变 RAMD。使用基于鲍曼不动杆菌推定胆固醇氧化酶 ORF 的基因特异性引物,分别在 pGEM®-T 和 pET-28a(+) 载体中克隆和表达胆固醇氧化酶基因菌株 AB030(GenBank [gb] 基因座标签:IX87_05230)。获得的核苷酸序列 (1671 bp, gb: MK575469.2),翻译成称为 choxAB (556 个氨基酸) 的蛋白质,在 1 m m IPTG 诱导的大肠杆菌中作为包涵体 (IB) (MW ~ 62 kDa) 过表达BL21 (DE3) 罗塞塔细胞。优化的表达条件(1 m m IPTG 与 2% [v/v] 甘油和在室温下)产生了 0.45 U·mL -1的可溶性活性 choxAB,增强了 56.25 倍。使用具有比活性(0.054 U·mg -1)、产率 (8.1%) 和倍数纯化 (11.69)。毛细管等电聚焦表明 choxAB 的 pI 为 8.77。LC-MS/MS 证实了 IB (62 kDa),其中 82.6% 的覆盖序列是鲍曼不动杆菌胆固醇氧化酶 (UniProtKB: A0A0E1FG24) 独有的。使用 LOMETS webtool 和Streptomyces sp. 的胆固醇氧化酶模板预测 choxAB 的 3D 结构。SA-COO(PDB:2GEW)。预测的二级结构包括 18 个 α-螺旋和 12 个 β-链、一个预测的催化三联体(E 220、H 380和 N 514)和一个保守的 FAD 结合序列(GSGFGGSVSACRLTEKG)。未来的研究应该考虑融合到溶解标签和转换到表达宿主毕赤酵母以减少 IB 的形成。
更新日期:2021-09-01
中文翻译:

不动杆菌属的胆固醇氧化酶的克隆、表达和计算机结构建模。大肠杆菌中的 RAMD 菌株
胆固醇氧化酶 (CHOX) 是黄素腺嘌呤二核苷酸依赖性氧化还原酶,具有一系列生物技术应用。仍然迫切需要确定新的 CHOX 家族成员以满足全球酶市场的需求。在这里,我们报告了不动杆菌属的胆固醇氧化酶的克隆、异源表达和结构建模。应变 RAMD。使用基于鲍曼不动杆菌推定胆固醇氧化酶 ORF 的基因特异性引物,分别在 pGEM®-T 和 pET-28a(+) 载体中克隆和表达胆固醇氧化酶基因菌株 AB030(GenBank [gb] 基因座标签:IX87_05230)。获得的核苷酸序列 (1671 bp, gb: MK575469.2),翻译成称为 choxAB (556 个氨基酸) 的蛋白质,在 1 m m IPTG 诱导的大肠杆菌中作为包涵体 (IB) (MW ~ 62 kDa) 过表达BL21 (DE3) 罗塞塔细胞。优化的表达条件(1 m m IPTG 与 2% [v/v] 甘油和在室温下)产生了 0.45 U·mL -1的可溶性活性 choxAB,增强了 56.25 倍。使用具有比活性(0.054 U·mg -1)、产率 (8.1%) 和倍数纯化 (11.69)。毛细管等电聚焦表明 choxAB 的 pI 为 8.77。LC-MS/MS 证实了 IB (62 kDa),其中 82.6% 的覆盖序列是鲍曼不动杆菌胆固醇氧化酶 (UniProtKB: A0A0E1FG24) 独有的。使用 LOMETS webtool 和Streptomyces sp. 的胆固醇氧化酶模板预测 choxAB 的 3D 结构。SA-COO(PDB:2GEW)。预测的二级结构包括 18 个 α-螺旋和 12 个 β-链、一个预测的催化三联体(E 220、H 380和 N 514)和一个保守的 FAD 结合序列(GSGFGGSVSACRLTEKG)。未来的研究应该考虑融合到溶解标签和转换到表达宿主毕赤酵母以减少 IB 的形成。



























 京公网安备 11010802027423号
京公网安备 11010802027423号