Molecular Cell ( IF 16.0 ) Pub Date : 2021-06-29 , DOI: 10.1016/j.molcel.2021.06.006 Sergei Rudnizky 1 , Hadeel Khamis 2 , Yuval Ginosar 1 , Efrat Goren 1 , Philippa Melamed 3 , Ariel Kaplan 3
|
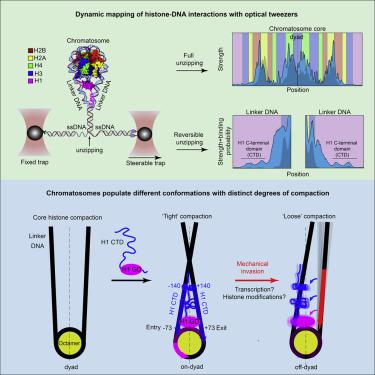
Chromatosomes play a fundamental role in chromatin regulation, but a detailed understanding of their structure is lacking, partially due to their complex dynamics. Using single-molecule DNA unzipping with optical tweezers, we reveal that linker histone interactions with DNA are remarkably extended, with the C-terminal domain binding both DNA linkers as far as approximately ±140 bp from the dyad. In addition to a symmetrical compaction of the nucleosome core governed by globular domain contacts at the dyad, the C-terminal domain compacts the nucleosome’s entry and exit. These interactions are dynamic, exhibit rapid binding and dissociation, are sensitive to phosphorylation of a specific residue, and are crucial to determining the symmetry of the chromatosome’s core. Extensive unzipping of the linker DNA, which mimics its invasion by motor proteins, shifts H1 into an asymmetric, off-dyad configuration and triggers nucleosome decompaction, highlighting the plasticity of the chromatosome structure and its potential regulatory role.
中文翻译:

扩展和动态接头组蛋白-DNA 相互作用控制染色体压实
染色体在染色质调控中发挥着重要作用,但缺乏对其结构的详细了解,部分原因是其复杂的动力学。使用光镊解压单分子 DNA,我们发现接头组蛋白与 DNA 的相互作用显着扩展,C 端域结合两个 DNA 接头,距离二元组约 ±140 bp。除了由二元体处的球状结构域接触控制的核小体核心的对称压缩之外,C 端结构域还压缩了核小体的进入和退出。这些相互作用是动态的,表现出快速结合和解离,对特定残基的磷酸化敏感,并且对于确定染色体核心的对称性至关重要。链接器 DNA 的广泛解链,模拟运动蛋白的入侵,



























 京公网安备 11010802027423号
京公网安备 11010802027423号