Journal of Structural Biology ( IF 3 ) Pub Date : 2021-06-27 , DOI: 10.1016/j.jsb.2021.107765 Darrell W Cockburn 1 , Ryan Kibler 1 , Haley A Brown 1 , Rebecca Duvall 1 , Sarah Moraïs 2 , Edward Bayer 3 , Nicole M Koropatkin 1
|
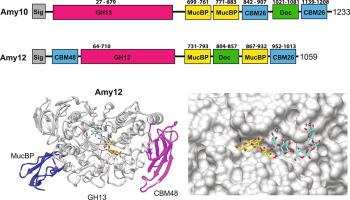
Pullulanases are glycoside hydrolase family 13 (GH13) enzymes that target α1,6 glucosidic linkages within starch and aid in the degradation of the α1,4- and α1,6- linked glucans pullulan, glycogen and amylopectin. The human gut bacterium Ruminococcus bromii synthesizes two extracellular pullulanases, Amy10 and Amy12, that are incorporated into the multiprotein amylosome complex that enables the digestion of granular resistant starch from the diet. Here we provide a comparative biochemical analysis of these pullulanases and the x-ray crystal structures of the wild type and the nucleophile mutant D392A of Amy12 complexed with maltoheptaose and 63-α-D glucosyl-maltotriose. While Amy10 displays higher catalytic efficiency on pullulan and cleaves only α1,6 linkages, Amy12 has some activity on α1,4 linkages suggesting that these enzymes are not redundant within the amylosome. Our structures of Amy12 include a mucin-binding protein (MucBP) domain that follows the C-domain of the GH13 fold, an atypical feature of these enzymes. The wild type Amy12 structure with maltoheptaose captured two oligosaccharides in the active site arranged as expected following catalysis of an α1,6 branch point in amylopectin. The nucleophile mutant D392A complexed with maltoheptaose or 63-α-D glucosyl-maltotriose captured β-glucose at the reducing end in the −1 subsite, facilitated by the truncation of the active site aspartate and stabilized by stacking with Y279. The core interface between the co-crystallized ligands and Amy12 occurs within the −2 through + 1 subsites, which may allow for flexible recognition of α1,6 linkages within a variety of starch structures.
中文翻译:

溴瘤胃球菌淀粉体支链淀粉酶的结构和底物识别
支链淀粉酶是糖苷水解酶家族 13 (GH13) 酶,其靶向淀粉内的 α1,6 糖苷键并有助于降解 α1,4- 和 α1,6- 连接的葡聚糖支链淀粉、糖原和支链淀粉。人类肠道细菌Ruminococcus bromii合成两种细胞外支链淀粉酶 Amy10 和 Amy12,它们被整合到多蛋白淀粉体复合物中,从而能够从饮食中消化颗粒状抗性淀粉。在这里,我们提供了这些支链淀粉酶的比较生化分析以及野生型的 X 射线晶体结构和 Amy12 与麦芽七糖和 6 3复合的亲核试剂突变体 D392A-α-D 葡糖基-麦芽三糖。虽然 Amy10 对支链淀粉表现出更高的催化效率并且仅切割 α1,6 键,但 Amy12 对 α1,4 键具有一定的活性,这表明这些酶在淀粉体中不是多余的。我们的 Amy12 结构包括一个粘蛋白结合蛋白 (MucBP) 结构域,该结构域位于 GH13 折叠的 C 结构域之后,这是这些酶的非典型特征。具有麦芽七糖的野生型 Amy12 结构在支链淀粉中的 α1,6 分支点催化后按预期排列的活性位点中捕获了两个寡糖。与麦芽七糖或 6 3复合的亲核试剂突变体 D392A-α-D 葡糖基-麦芽三糖在 -1 亚位点的还原端捕获 β-葡萄糖,通过截断活性位点天冬氨酸促进并通过与 Y279 堆叠来稳定。共结晶配体和 Amy12 之间的核心界面出现在 -2 到 + 1 个亚位点内,这可以灵活识别各种淀粉结构中的 α1,6 键。



























 京公网安备 11010802027423号
京公网安备 11010802027423号