Water Research ( IF 12.8 ) Pub Date : 2021-06-17 , DOI: 10.1016/j.watres.2021.117369 Nicole Acosta 1 , María A Bautista 2 , Jordan Hollman 3 , Janine McCalder 4 , Alexander Buchner Beaudet 2 , Lawrence Man 2 , Barbara J Waddell 1 , Jianwei Chen 2 , Carmen Li 2 , Darina Kuzma 5 , Srijak Bhatnagar 2 , Jenine Leal 6 , Jon Meddings 7 , Jia Hu 8 , Jason L Cabaj 9 , Norma J Ruecker 10 , Christopher Naugler 11 , Dylan R Pillai 12 , Gopal Achari 13 , M Cathryn Ryan 14 , John M Conly 15 , Kevin Frankowski 5 , Casey Rj Hubert 2 , Michael D Parkins 16
|
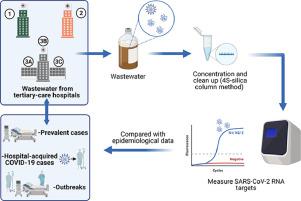
SARS-CoV-2 has been detected in wastewater and its abundance correlated with community COVID-19 cases, hospitalizations and deaths. We sought to use wastewater-based detection of SARS-CoV-2 to assess the epidemiology of SARS-CoV-2 in hospitals. Between August and December 2020, twice-weekly wastewater samples from three tertiary-care hospitals (totaling >2100 dedicated inpatient beds) were collected. Hospital-1 and Hospital-2 could be captured with a single sampling point whereas Hospital-3 required three separate monitoring sites. Wastewater samples were concentrated and cleaned using the 4S-silica column method and assessed for SARS-CoV-2 gene-targets (N1, N2 and E) and controls using RT-qPCR. Wastewater SARS-CoV-2 as measured by quantification cycle (Cq), genome copies and genomes normalized to the fecal biomarker PMMoV were compared to the total daily number of patients hospitalized with active COVID-19, confirmed cases of hospital-acquired infection, and the occurrence of unit-specific outbreaks. Of 165 wastewater samples collected, 159 (96%) were assayable. The N1-gene from SARS-CoV-2 was detected in 64.1% of samples, N2 in 49.7% and E in 10%. N1 and N2 in wastewater increased over time both in terms of the amount of detectable virus and the proportion of samples that were positive, consistent with increasing hospitalizations at those sites with single monitoring points (Pearson's r=0.679, P<0.0001, Pearson's r=0.799, P<0.0001, respectively). Despite increasing hospitalizations through the study period, nosocomial-acquired cases of COVID-19 (Pearson's r =0.389, P<0.001) and unit-specific outbreaks were discernable with significant increases in detectable SARS-CoV-2 N1-RNA (median 112 copies/ml) versus outbreak-free periods (0 copies/ml; P<0.0001). Wastewater-based monitoring of SARS-CoV-2 represents a promising tool for SARS-CoV-2 passive surveillance and case identification, containment, and mitigation in acute- care medical facilities.
中文翻译:

一项调查三级医院废水中 SARS-CoV-2 的多中心研究。病毒负担与住院病例的增加以及医院相关的传播和暴发有关
已在废水中检测到 SARS-CoV-2,其丰度与社区 COVID-19 病例、住院和死亡相关。我们试图使用基于废水的 SARS-CoV-2 检测来评估医院中 SARS-CoV-2 的流行病学。2020 年 8 月至 2020 年 12 月期间,从三家三级医院(总计超过 2100 张专用住院床位)收集了每周两次的废水样本。Hospital-1 和 Hospital-2 可以通过单个采样点捕获,而 Hospital-3 需要三个独立的监测点。使用 4S-硅胶柱法浓缩和清洁废水样本,并使用 RT-qPCR 评估 SARS-CoV-2 基因靶标(N1、N2 和 E)和对照。通过量化循环 (Cq) 测量的废水 SARS-CoV-2,将基因组拷贝和标准化为粪便生物标志物 PMMoV 的基因组与每天住院的活跃 COVID-19 患者总数、确诊的医院获得性感染病例以及特定单位暴发的发生率进行比较。在收集到的 165 个废水样本中,有 159 个 (96%) 是可检测的。在 64.1% 的样本中检测到来自 SARS-CoV-2 的 N1 基因,在 49.7% 的样本中检测到 N2,在 10% 的样本中检测到 E。随着时间的推移,废水中的 N1 和 N2 在可检测病毒的数量和阳性样本比例方面均有所增加,这与那些具有单一监测点的地点的住院人数增加一致(Pearson's r=0.679,P<0.0001,Pearson's r= 0.799,P<0.0001)。尽管在整个研究期间住院人数有所增加,但院内获得性 COVID-19 病例(Pearson's r =0.389,P<0. 001)和特定单位的爆发是可识别的,可检测到的 SARS-CoV-2 N1-RNA(中位数 112 拷贝/毫升)与无爆发期(0 拷贝/毫升;P<0.0001)相比显着增加。基于废水的 SARS-CoV-2 监测是一种很有前途的工具,可用于急救医疗设施中的 SARS-CoV-2 被动监测和病例识别、遏制和缓解。

























 京公网安备 11010802027423号
京公网安备 11010802027423号