当前位置:
X-MOL 学术
›
Immunol. Cell Biol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
SARS-CoV-2-specific CD8+ T-cell responses and TCR signatures in the context of a prominent HLA-A*24:02 allomorph
Immunology and Cell Biology ( IF 4 ) Pub Date : 2021-06-04 , DOI: 10.1111/imcb.12482 Louise C Rowntree 1 , Jan Petersen 2, 3 , Jennifer A Juno 1 , Priyanka Chaurasia 2 , Kathleen Wragg 1 , Marios Koutsakos 1 , Luca Hensen 1 , Adam K Wheatley 1, 4 , Stephen J Kent 1, 4, 5 , Jamie Rossjohn 2, 3, 6 , Katherine Kedzierska 1 , Thi HO Nguyen 1
Immunology and Cell Biology ( IF 4 ) Pub Date : 2021-06-04 , DOI: 10.1111/imcb.12482 Louise C Rowntree 1 , Jan Petersen 2, 3 , Jennifer A Juno 1 , Priyanka Chaurasia 2 , Kathleen Wragg 1 , Marios Koutsakos 1 , Luca Hensen 1 , Adam K Wheatley 1, 4 , Stephen J Kent 1, 4, 5 , Jamie Rossjohn 2, 3, 6 , Katherine Kedzierska 1 , Thi HO Nguyen 1
Affiliation
|
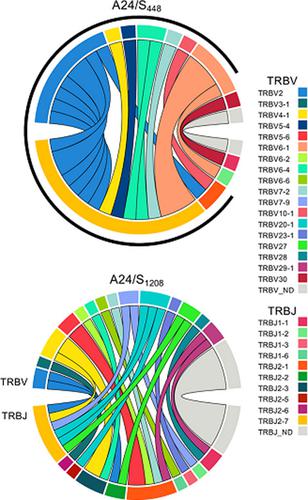
In-depth understanding of human T-cell-mediated immunity in coronavirus disease 2019 (COVID-19) is needed if we are to optimize vaccine strategies and immunotherapies. Identification of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) T-cell epitopes and generation of peptide–human leukocyte antigen (peptide–HLA) tetramers facilitate direct ex vivo analyses of SARS-CoV-2-specific T cells and their T-cell receptor (TCR) repertoires. We utilized a combination of peptide prediction and in vitro peptide stimulation to validate novel SARS-CoV-2 epitopes restricted by HLA-A*24:02, one of the most prominent HLA class I alleles, especially in Indigenous and Asian populations. Of the peptides screened, three spike-derived peptides generated CD8+IFNγ+ responses above background, S1208–1216 (QYIKWPWYI), S448–456 (NYNYLYRLF) and S193–201 (VFKNIDGYF), with S1208 generating immunodominant CD8+IFNγ+ responses. Using peptide–HLA-I tetramers, we performed direct ex vivo tetramer enrichment for HLA-A*24:02-restricted CD8+ T cells in COVID-19 patients and prepandemic controls. The precursor frequencies for HLA-A*24:02-restricted epitopes were within the range previously observed for other SARS-CoV-2 epitopes for both COVID-19 patients and prepandemic individuals. Naïve A24/SARS-CoV-2-specific CD8+ T cells increased nearly 7.5-fold above the average precursor frequency during COVID-19, gaining effector and memory phenotypes. Ex vivo single-cell analyses of TCRαβ repertoires found that the A24/S448+CD8+ T-cell TCRαβ repertoire was driven by a common TCRβ chain motif, whereas the A24/S1208+CD8+ TCRαβ repertoire was diverse across COVID-19 patients. Our study provides an in depth characterization and important insights into SARS-CoV-2-specific CD8+ T-cell responses associated with a prominent HLA-A*24:02 allomorph. This contributes to our knowledge on adaptive immune responses during primary COVID-19 and could be exploited in vaccine or immunotherapeutic approaches.
中文翻译:

在突出的 HLA-A*24:02 异形体背景下,SARS-CoV-2 特异性 CD8+ T 细胞反应和 TCR 特征
如果我们要优化疫苗策略和免疫疗法,就需要深入了解 2019 年冠状病毒病 (COVID-19) 中人类 T 细胞介导的免疫。严重急性呼吸综合征冠状病毒 2 (SARS-CoV-2) T 细胞表位的鉴定和肽-人类白细胞抗原 (肽-HLA) 四聚体的产生促进了对 SARS-CoV-2 特异性 T 细胞及其特异性的直接离体分析T 细胞受体 (TCR) 库。我们结合肽预测和体外肽刺激来验证受 HLA-A*24:02 限制的新型 SARS-CoV-2 表位,HLA-A*24:02 是最突出的 HLA I 类等位基因之一,尤其是在土著和亚洲人群中。在筛选的肽中,三种尖峰衍生肽产生 CD8 + IFNγ+高于背景的反应,S 1208-1216 (QYIKWPWYI)、S 448-456 (NYNYLYRLF) 和 S 193-201 (VFKNIDGYF),S 1208产生免疫显性 CD8 + IFNγ +反应。使用肽-HLA-I 四聚体,我们对 COVID-19 患者和大流行前对照中的HLA-A*24:02 限制性 CD8 + T 细胞进行了直接的离体四聚体富集。HLA-A*24:02 限制性表位的前体频率在先前观察到的 COVID-19 患者和大流行前个体的其他 SARS-CoV-2 表位的范围内。Naïve A24/SARS-CoV-2 特异性 CD8 +在 COVID-19 期间,T 细胞比平均前体频率增加了近 7.5 倍,获得了效应和记忆表型。TCRαβ 库的离体单细胞分析发现 A24/S 448 + CD8 + T 细胞 TCRαβ 库是由一个常见的 TCRβ 链基序驱动的,而 A24/S 1208 + CD8 + TCRαβ 库在 COVID-19 中是多种多样的耐心。我们的研究提供了对 SARS-CoV-2 特异性 CD8 + 的深入表征和重要见解与突出的 HLA-A*24:02 异形体相关的 T 细胞反应。这有助于我们了解初级 COVID-19 期间的适应性免疫反应,并可用于疫苗或免疫治疗方法。
更新日期:2021-06-04
中文翻译:

在突出的 HLA-A*24:02 异形体背景下,SARS-CoV-2 特异性 CD8+ T 细胞反应和 TCR 特征
如果我们要优化疫苗策略和免疫疗法,就需要深入了解 2019 年冠状病毒病 (COVID-19) 中人类 T 细胞介导的免疫。严重急性呼吸综合征冠状病毒 2 (SARS-CoV-2) T 细胞表位的鉴定和肽-人类白细胞抗原 (肽-HLA) 四聚体的产生促进了对 SARS-CoV-2 特异性 T 细胞及其特异性的直接离体分析T 细胞受体 (TCR) 库。我们结合肽预测和体外肽刺激来验证受 HLA-A*24:02 限制的新型 SARS-CoV-2 表位,HLA-A*24:02 是最突出的 HLA I 类等位基因之一,尤其是在土著和亚洲人群中。在筛选的肽中,三种尖峰衍生肽产生 CD8 + IFNγ+高于背景的反应,S 1208-1216 (QYIKWPWYI)、S 448-456 (NYNYLYRLF) 和 S 193-201 (VFKNIDGYF),S 1208产生免疫显性 CD8 + IFNγ +反应。使用肽-HLA-I 四聚体,我们对 COVID-19 患者和大流行前对照中的HLA-A*24:02 限制性 CD8 + T 细胞进行了直接的离体四聚体富集。HLA-A*24:02 限制性表位的前体频率在先前观察到的 COVID-19 患者和大流行前个体的其他 SARS-CoV-2 表位的范围内。Naïve A24/SARS-CoV-2 特异性 CD8 +在 COVID-19 期间,T 细胞比平均前体频率增加了近 7.5 倍,获得了效应和记忆表型。TCRαβ 库的离体单细胞分析发现 A24/S 448 + CD8 + T 细胞 TCRαβ 库是由一个常见的 TCRβ 链基序驱动的,而 A24/S 1208 + CD8 + TCRαβ 库在 COVID-19 中是多种多样的耐心。我们的研究提供了对 SARS-CoV-2 特异性 CD8 + 的深入表征和重要见解与突出的 HLA-A*24:02 异形体相关的 T 细胞反应。这有助于我们了解初级 COVID-19 期间的适应性免疫反应,并可用于疫苗或免疫治疗方法。


























 京公网安备 11010802027423号
京公网安备 11010802027423号