Current Genomics ( IF 2.6 ) Pub Date : 2021-01-31 , DOI: 10.2174/1389202922666210301084151 Jeaneth Machicao 1 , Francesco Craighero 1 , Davide Maspero 1 , Fabrizio Angaroni 1 , Chiara Damiani 1 , Alex Graudenzi 1 , Marco Antoniotti 1 , Odemir M Bruno 1
|
Background: The increasing availability of omics data collected from patients affected by severe pathologies, such as cancer, is fostering the development of data science methods for their analysis.
Introduction: The combination of data integration and machine learning approaches can provide new powerful instruments to tackle the complexity of cancer development and deliver effective diagnostic and prognostic strategies.
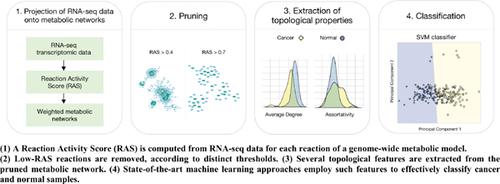
Methods: We explore the possibility of exploiting the topological properties of sample-specific metabolic networks as features in a supervised classification task. Such networks are obtained by projecting transcriptomic data from RNA-seq experiments on genome-wide metabolic models to define weighted networks modeling the overall metabolic activity of a given sample.
Results: We show the classification results on a labeled breast cancer dataset from the TCGA database, including 210 samples (cancer vs. normal). In particular, we investigate how the performance is affected by a threshold-based pruning of the networks by comparing Artificial Neural Networks, Support Vector Machines and Random Forests. Interestingly, the best classification performance is achieved within a small threshold range for all methods, suggesting that it might represent an effective choice to recover useful information while filtering out noise from data. Overall, the best accuracy is achieved with SVMs, which exhibit performances similar to those obtained when gene expression profiles are used as features.
Conclusion: These findings demonstrate that the topological properties of sample-specific metabolic networks are effective in classifying cancer and normal samples, suggesting that useful information can be extracted from a relatively limited number of features.
中文翻译:

关于利用代谢网络的拓扑特征对癌症样本进行分类
背景:从受癌症等严重疾病影响的患者身上收集的组学数据越来越多,正在促进数据科学方法的发展,以进行分析。
简介:数据集成和机器学习方法的结合可以提供新的强大工具来解决癌症发展的复杂性并提供有效的诊断和预后策略。
方法:我们探索利用特定样本代谢网络的拓扑特性作为监督分类任务中的特征的可能性。此类网络是通过将 RNA-seq 实验的转录组数据投影到全基因组代谢模型上来定义加权网络来模拟给定样本的整体代谢活动而获得的。
结果:我们展示了 TCGA 数据库中标记的乳腺癌数据集的分类结果,包括 210 个样本(癌症与正常)。特别是,我们通过比较人工神经网络、支持向量机和随机森林来研究基于阈值的网络修剪如何影响性能。有趣的是,所有方法的最佳分类性能都是在一个小的阈值范围内实现的,这表明它可能代表了一种在从数据中滤除噪声的同时恢复有用信息的有效选择。总体而言,使用 SVM 实现了最佳精度,其表现出与使用基因表达谱作为特征时获得的性能相似的性能。
结论:这些发现表明,样本特异性代谢网络的拓扑特性在分类癌症和正常样本方面是有效的,这表明可以从相对有限的特征中提取有用的信息。


























 京公网安备 11010802027423号
京公网安备 11010802027423号