Current Genomics ( IF 2.6 ) Pub Date : 2021-01-31 , DOI: 10.2174/1389202922666210219110831 Maddalena Dilucca 1 , Giulio Cimini 2 , Andrea Giansanti 3
|
Background: Protein-protein interaction (PPI) networks are the backbone of all processes in living cells. In this work, we relate conservation, essentiality and functional repertoire of a gene to the connectivity k (i.e. the number of interactions, links) of the corresponding protein in the PPI network.
Methods: On a set of 42 bacterial genomes of different sizes, and with reasonably separated evolutionary trajectories, we investigate three issues: i) whether the distribution of connectivities changes between PPI subnetworks of essential and nonessential genes; ii) how gene conservation, measured both by the evolutionary retention index (ERI) and by evolutionary pressures, is related to the connectivity of the corresponding protein; iii) how PPI connectivities are modulated by evolutionary and functional relationships, as represented by the Clusters of Orthologous Genes (COGs).
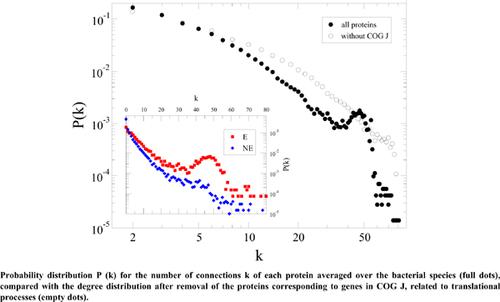
Results: We show that conservation, essentiality and functional specialisation of genes constrain the connectivity of the corresponding proteins in bacterial PPI networks. In particular, we isolated a core of highly connected proteins (connectivities k≥40), which is ubiquitous among the species considered here, though mostly visible in the degree distributions of bacteria with small genomes (less than 1000 genes).
Conclusion: The genes that support this highly connected core are conserved, essential and, in most cases, belong to the COG cluster J, related to ribosomal functions and the processing of genetic information.
中文翻译:

细菌蛋白质相互作用网络:连接性受基因保守性、本质和功能支配
背景:蛋白质-蛋白质相互作用 (PPI) 网络是活细胞中所有过程的支柱。在这项工作中,我们将基因的保守性、必要性和功能库与 PPI 网络中相应蛋白质的连接性 k(即相互作用、链接的数量)联系起来。
方法:在一组 42 个不同大小的细菌基因组上,具有合理分离的进化轨迹,我们研究了三个问题:i)必需基因和非必需基因的 PPI 子网络之间的连通性分布是否发生变化;ii) 通过进化保留指数 (ERI) 和进化压力测量的基因保守性如何与相应蛋白质的连接性相关;iii) PPI 连接如何被进化和功能关系调节,如直系同源基因簇 (COG) 所代表。
结果:我们表明,基因的保守性、必要性和功能特化限制了细菌 PPI 网络中相应蛋白质的连接性。特别是,我们分离了一个高度连接的蛋白质核心(连接性 k≥40),这在此处考虑的物种中无处不在,尽管在具有小基因组(少于 1000 个基因)的细菌的程度分布中大部分可见。
结论:支持这种高度连接核心的基因是保守的、必不可少的,并且在大多数情况下属于与核糖体功能和遗传信息处理有关的 COG 簇 J。


























 京公网安备 11010802027423号
京公网安备 11010802027423号