Protein & Peptide Letters ( IF 1.6 ) Pub Date : 2021-08-31 , DOI: 10.2174/0929866528666210405160131 Sasikumar Sabna 1 , Dev Vrat Kamboj 1 , Ravi Bhushan Kumar 1 , Prabhakar Babele 1 , Sakshi Rajoria 1 , Mahendra Kumar Gupta 2 , Syed Imteyaz Alam 1
|
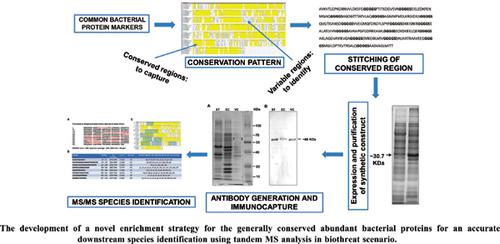
Background: Some pathogenic bacteria can be potentially used for nefarious applications in the event of bioterrorism or biowarfare. Accurate identification of biological agent from clinical and diverse environmental matrices is of paramount importance for implementation of medical countermeasures and biothreat mitigation.
Objective: A novel methodology is reported here for the development of a novel enrichment strategy for the generally conserved abundant bacterial proteins for an accurate downstream species identification using tandem MS analysis in biothreat scenario.
Methods: Conserved regions in the common bacterial protein markers were analyzed using bioinformatic tools and stitched for a possible generic immuno-capture for an intended downstream MS/MS analysis. Phylogenetic analysis of selected proteins was carried out and synthetic constructs were generated for the expression of conserved stitched regions of 60 kDa chaperonin GroEL. Hyper-immune serum was raised against recombinant synthetic GroEL protein.
Results: The conserved regions of common bacterial proteins were stitched for a possible generic immuno-capture and subsequent specific identification by tandem MS using variable regions of the molecule. Phylogenetic analysis of selected proteins was carried out and synthetic constructs were generated for the expression of conserved stitched regions of GroEL. In a proof-of-concept study, hyper-immune serum raised against recombinant synthetic GroEL protein exhibited reactivity with ~60 KDa proteins from the cell lysates of three bacterial species tested.
Conclusion: The envisaged methodology can lead to the development of a novel enrichment strategy for the abundant bacterial proteins from complex environmental matrices for the downstream species identification with increased sensitivity and substantially reduce the time-to-result.
中文翻译:

从不同细菌选择剂中富集蛋白质生物标志物的策略
背景:在发生生物恐怖主义或生物战的情况下,某些病原菌可能被用于恶意应用。从临床和各种环境基质中准确识别生物制剂对于实施医疗对策和减轻生物威胁至关重要。
目标:这里报告了一种新方法,用于为普遍保守的丰富细菌蛋白质开发新的富集策略,以便在生物威胁场景中使用串联 MS 分析准确识别下游物种。
方法:使用生物信息学工具分析常见细菌蛋白质标记中的保守区域,并缝合可能的通用免疫捕获,用于预期的下游 MS/MS 分析。对选定的蛋白质进行了系统发育分析,并生成了用于表达 60 kDa 伴侣蛋白 GroEL 的保守缝合区域的合成构建体。针对重组合成 GroEL 蛋白产生超免疫血清。
结果:常见细菌蛋白质的保守区域被缝合,用于可能的通用免疫捕获和随后使用分子可变区的串联 MS 进行特异性识别。对选定的蛋白质进行了系统发育分析,并生成了用于表达 GroEL 的保守缝合区域的合成构建体。在一项概念验证研究中,针对重组合成 GroEL 蛋白产生的超免疫血清表现出与来自三种测试细菌物种细胞裂解物的约 60 KDa 蛋白的反应性。
结论:设想的方法可以导致开发一种新的富集策略,从复杂的环境基质中提取丰富的细菌蛋白质,用于下游物种鉴定,提高灵敏度并大大缩短获得结果的时间。



























 京公网安备 11010802027423号
京公网安备 11010802027423号