Current Bioinformatics ( IF 4 ) Pub Date : 2021-01-31 , DOI: 10.2174/1574893615999200601122840 Ashish Kumar Sharma 1 , Rajeev Srivastava 1
|
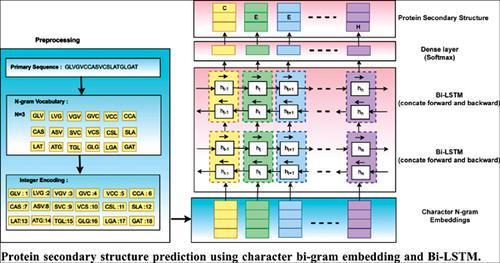
Background: Protein secondary structure is vital to predicting the tertiary structure, which is essential in deciding protein function and drug designing. Therefore, there is a high requirement of computational methods to predict secondary structure from their primary sequence. Protein primary sequences represented as a linear combination of twenty amino acid characters and contain the contextual information for secondary structure prediction.
Objective and Methods: Protein secondary structure predicted from their primary sequences using a deep recurrent neural network. Protein secondary structure depends on local and long-range residues in primary sequences. In the proposed work, the local contextual information of amino acid residues captures with character n-gram. A dense embedding vector represents this local contextual information. Furthermore, the bidirectional long short-term memory (Bi-LSTM) model is used to capture the long-range contexts by extracting the past and future residues information in primary sequences.
Results: The proposed deep recurrent architecture is evaluated for its efficacy for datasets, namely ss.txt, RS126, and CASP9. The model shows the Q3 accuracies of 88.45%, 83.48%, and 86.69% for ss.txt, RS126, and CASP9, respectively. The performance of the proposed model is also compared with other state-of-the-art methods available in the literature.
Conclusion: After a comparative analysis, it was observed that the proposed model is performing better in comparison to state-of-art methods.
中文翻译:

使用字符二聚体嵌入和Bi-LSTM预测蛋白质二级结构
背景:蛋白质二级结构对于预测三级结构至关重要,这对决定蛋白质功能和药物设计至关重要。因此,对从其一级序列预测二级结构的计算方法提出了很高的要求。蛋白质一级序列表示为二十个氨基酸特征的线性组合,并包含用于二级结构预测的上下文信息。
目的和方法:使用深度递归神经网络从其一级序列预测蛋白质二级结构。蛋白质二级结构取决于一级序列中的局部和远距离残基。在拟议的工作中,氨基酸残基的局部上下文信息以字符n-gram捕获。密集的嵌入向量表示此局部上下文信息。此外,双向长期短期记忆(Bi-LSTM)模型用于通过提取一级序列中过去和将来的残基信息来捕获远程上下文。
结果:评估了拟议的深度递归体系结构对数据集ss.txt,RS126和CASP9的功效。该模型显示ss.txt,RS126和CASP9的Q3准确性分别为88.45%,83.48%和86.69%。提出的模型的性能也与文献中提供的其他最新方法进行了比较。
结论:经过比较分析,发现与最新方法相比,该模型的性能更好。



























 京公网安备 11010802027423号
京公网安备 11010802027423号