Journal of Structural Biology ( IF 3 ) Pub Date : 2021-04-06 , DOI: 10.1016/j.jsb.2021.107736 Kent R Thurber 1 , Yi Yin 2 , Robert Tycko 1
|
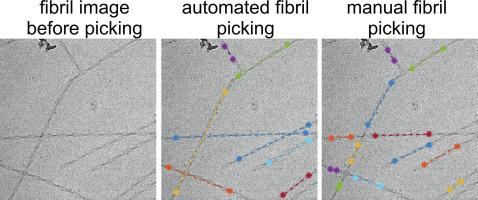
Cryogenic electron microscopy (cryo-EM) is an important tool for determining the molecular structure of proteins and protein assemblies, including helical assemblies such as amyloid fibrils. In reconstruction of amyloid fibril structures from cryo-EM images, an important early step is the selection of fibril locations. This fibril picking step is typically done by hand, a tedious process when thousands of images need to be analyzed. Here we present a computer program called FibrilFinder that identifies the locations and directions of fibril segments in cryo-EM images, by using the properties that the fibrils should be linear objects and have widths within a specified range. The program outputs the fibril locations in text files compatible with the RELION density reconstruction program. After RELION is used to extract the particle image boxes contained in the fibril segments identified by FibrilFinder, a second program called FibrilFixer removes boxes that contain more than one fibril, for instance because two fibrils cross each other. As concrete and realistic examples, we describe the application of the two programs to cryo-EM images of two different amyloid fibrils, namely 40-residue amyloid-β fibrils derived from human brain tissue by seeded growth and fibrils formed by the C-terminal half of the low-complexity domain of the RNA-binding protein FUS. Both examples of amyloid fibrils can be picked from cryo-EM images using the same set of FibrilFinder and FibrilFixer parameters, showing that this software does not require re-optimization for each sample. A set of 1337 cryo-EM images was analyzed in 17 min with one multi-core computer. The new fibril picking software should enable the rapid analysis and comparison of more helical structures using cryo-EM, and perhaps serve as part of the greater automation of the entire structure determination process.
中文翻译:

使用 RELION 从低温 EM 图像中自动挑选淀粉样蛋白原纤维用于螺旋重建
低温电子显微镜 (cryo-EM) 是确定蛋白质和蛋白质组装体(包括淀粉样蛋白原纤维等螺旋组装体)分子结构的重要工具。在从冷冻电镜图像重建淀粉样蛋白原纤维结构时,一个重要的早期步骤是选择原纤维位置。这个纤维拾取步骤通常是手工完成的,当需要分析数千张图像时,这是一个乏味的过程。在这里,我们提出了一个名为 FibrilFinder 的计算机程序,它通过使用原纤维应该是线性物体并且具有指定范围内的宽度的特性来识别低温 EM 图像中原纤维片段的位置和方向。该程序在与 RELION 密度重建程序兼容的文本文件中输出原纤维位置。在使用 RELION 提取包含在由 FibrilFinder 识别的原纤维片段中的粒子图像框之后,名为 FibrilFixer 的第二个程序会删除包含多个原纤维的框,例如因为两条原纤维相互交叉。作为具体和现实的例子,我们描述了这两个程序在两种不同淀粉样蛋白原纤维的冷冻电镜图像中的应用,即通过种子生长从人脑组织中提取的 40 个残基淀粉样蛋白-β 原纤维和由 C 端半部分形成的原纤维RNA结合蛋白FUS的低复杂性结构域。淀粉样原纤维的两个例子都可以使用相同的 FibrilFinder 和 FibrilFixer 参数集从低温 EM 图像中挑选出来,这表明该软件不需要对每个样本进行重新优化。使用一台多核计算机在 17 分钟内分析了一组 1337 张冷冻电镜图像。新的原纤维拾取软件应该能够使用冷冻电镜快速分析和比较更多的螺旋结构,并可能作为整个结构确定过程更大自动化的一部分。



























 京公网安备 11010802027423号
京公网安备 11010802027423号