当前位置:
X-MOL 学术
›
Acc. Chem. Res.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Going the dHis-tance: Site-Directed Cu2+ Labeling of Proteins and Nucleic Acids
Accounts of Chemical Research ( IF 18.3 ) Pub Date : 2021-01-21 , DOI: 10.1021/acs.accounts.0c00761 Austin Gamble Jarvi 1 , Xiaowei Bogetti 1 , Kevin Singewald 1 , Shreya Ghosh 1 , Sunil Saxena 1
Accounts of Chemical Research ( IF 18.3 ) Pub Date : 2021-01-21 , DOI: 10.1021/acs.accounts.0c00761 Austin Gamble Jarvi 1 , Xiaowei Bogetti 1 , Kevin Singewald 1 , Shreya Ghosh 1 , Sunil Saxena 1
Affiliation
|
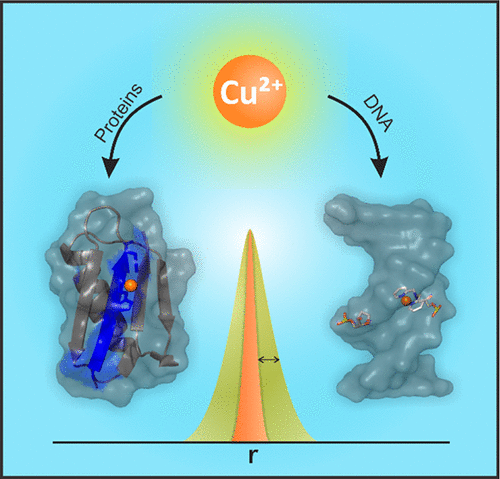
In this Account, we showcase site-directed Cu2+ labeling in proteins and DNA, which has opened new avenues for the measurement of the structure and dynamics of biomolecules using electron paramagnetic resonance (EPR) spectroscopy. In proteins, the spin label is assembled in situ from natural amino acid residues and a metal complex and requires no post-expression synthetic modification or purification procedures. The labeling scheme exploits a double histidine (dHis) motif, which utilizes endogenous or site-specifically mutated histidine residues to coordinate a Cu2+ complex. Pulsed EPR measurements on such Cu2+-labeled proteins potentially yield distance distributions that are up to 5 times narrower than the common protein spin label—the approach, thus, overcomes the inherent limitation of the current technology, which relies on a spin label with a highly flexible side chain. This labeling scheme provides a straightforward method that elucidates biophysical information that is costly, complicated, or simply inaccessible by traditional EPR labels. Examples include the direct measurement of protein backbone dynamics at β-sheet sites, which are largely inaccessible through traditional spin labels, and rigid Cu2+–Cu2+ distance measurements that enable higher precision in the analysis of protein conformations, conformational changes, interactions with other biomolecules, and the relative orientations of two labeled protein subunits. Likewise, a Cu2+ label has been developed for use in DNA, which is small, is nucleotide independent, and is positioned within the DNA helix. The placement of the Cu2+ label directly reports on the biologically relevant backbone distance. Additionally, for both of these labeling techniques, we have developed models for interpretation of the EPR distance information, primarily utilizing molecular dynamics (MD) simulations. Initial results using force fields developed for both protein and DNA labels have agreed with experimental results, which has been a major bottleneck for traditional spin labels. Looking ahead, we anticipate new combinations of MD and EPR to further our understanding of protein and DNA conformational changes, as well as working synergistically to investigate protein–DNA interactions.
中文翻译:

进行研究:蛋白质和核酸的定点Cu 2+标记
在此帐户中,我们展示了蛋白质和DNA中的定点Cu 2+标记,这为使用电子顺磁共振(EPR)光谱测量生物分子的结构和动力学开辟了新途径。在蛋白质中,旋转标记是从天然氨基酸残基和金属络合物原位组装的,不需要表达后的合成修饰或纯化程序。标记方案利用了双组氨酸(dHis)基序,该基序利用内源或位点特异性突变的组氨酸残基来协调Cu 2+复合物。对此类Cu 2+标记的蛋白质进行脉冲EPR测量可能会导致距离分布窄达5倍与普通的蛋白质旋转标签相比,该方法克服了当前技术的固有局限性,该技术依赖于具有高度柔性侧链的旋转标签。这种标记方案提供了一种简单的方法,可阐明传统EPR标签昂贵,复杂或根本无法获取的生物物理信息。例子包括直接测量β-折叠位点上的蛋白质骨架动力学(通过传统的旋转标记很难获得),以及刚性的Cu 2+ -Cu 2+距离测量,可在分析蛋白质构象,构象变化,相互作用中实现更高的精确度与其他生物分子,以及两个标记的蛋白质亚基的相对方向。同样,Cu 2+已经开发出用于DNA的小标签,该标签小,不依赖核苷酸并且位于DNA螺旋内。Cu 2+标签的位置直接报告了生物学相关的主链距离。此外,对于这两种标记技术,我们已经开发了用于解释EPR距离信息的模型,主要是利用分子动力学(MD)模拟。使用针对蛋白质和DNA标签开发的力场的初步结果与实验结果一致,这已成为传统旋转标签的主要瓶颈。展望未来,我们期待MD和EPR的新结合,以加深我们对蛋白质和DNA构象变化的了解,并协同研究蛋白质与DNA的相互作用。
更新日期:2021-03-16
中文翻译:

进行研究:蛋白质和核酸的定点Cu 2+标记
在此帐户中,我们展示了蛋白质和DNA中的定点Cu 2+标记,这为使用电子顺磁共振(EPR)光谱测量生物分子的结构和动力学开辟了新途径。在蛋白质中,旋转标记是从天然氨基酸残基和金属络合物原位组装的,不需要表达后的合成修饰或纯化程序。标记方案利用了双组氨酸(dHis)基序,该基序利用内源或位点特异性突变的组氨酸残基来协调Cu 2+复合物。对此类Cu 2+标记的蛋白质进行脉冲EPR测量可能会导致距离分布窄达5倍与普通的蛋白质旋转标签相比,该方法克服了当前技术的固有局限性,该技术依赖于具有高度柔性侧链的旋转标签。这种标记方案提供了一种简单的方法,可阐明传统EPR标签昂贵,复杂或根本无法获取的生物物理信息。例子包括直接测量β-折叠位点上的蛋白质骨架动力学(通过传统的旋转标记很难获得),以及刚性的Cu 2+ -Cu 2+距离测量,可在分析蛋白质构象,构象变化,相互作用中实现更高的精确度与其他生物分子,以及两个标记的蛋白质亚基的相对方向。同样,Cu 2+已经开发出用于DNA的小标签,该标签小,不依赖核苷酸并且位于DNA螺旋内。Cu 2+标签的位置直接报告了生物学相关的主链距离。此外,对于这两种标记技术,我们已经开发了用于解释EPR距离信息的模型,主要是利用分子动力学(MD)模拟。使用针对蛋白质和DNA标签开发的力场的初步结果与实验结果一致,这已成为传统旋转标签的主要瓶颈。展望未来,我们期待MD和EPR的新结合,以加深我们对蛋白质和DNA构象变化的了解,并协同研究蛋白质与DNA的相互作用。



























 京公网安备 11010802027423号
京公网安备 11010802027423号