Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
DNA modification and visualization on an origami-based enzyme nano-factory
Nanoscale ( IF 6.7 ) Pub Date : 2020-12-21 , DOI: 10.1039/d0nr07618j Elmar Weinhold 1, 2, 3, 4 , Banani Chakraborty 5, 6, 7, 8
Nanoscale ( IF 6.7 ) Pub Date : 2020-12-21 , DOI: 10.1039/d0nr07618j Elmar Weinhold 1, 2, 3, 4 , Banani Chakraborty 5, 6, 7, 8
Affiliation
|
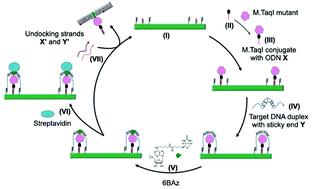
The past decade has seen enormous progress in DNA nanotechnology through the advent of DNA origami. Functionalizing the DNA origami for multiple applications is the recent focus of this field. Here we have constructed a novel DNA enzyme nano-factory, which modifies target DNA embedded on a DNA origami platform. The enzyme is programmed to reside in close proximity to the target DNA which enhances significantly the local concentration compared to solution-based DNA modification. To demonstrate this we have immobilized DNA methyltransferase M·TaqI next to the target DNA on the DNA origami and used this enzyme to sequence-specifically modify the target DNA with biotin using a cofactor analogue. Streptavidin binding to biotin is applied as a topographic marker to follow the machine cycle of this enzyme nano-factory using atomic force microscopy imaging. The nano-factory is demonstrated to be recyclable and holds the potential to be expanded to a multi-enzyme, multi-substrate operating system controlled by simple to complex molecules made of DNA, RNA or proteins.
中文翻译:

基于折纸的酶纳米工厂的DNA修饰和可视化
在过去的十年中,随着DNA折纸的出现,DNA纳米技术取得了巨大进步。使DNA折纸具有多种用途的功能是该领域的近期重点。在这里,我们构建了一个新型的DNA酶纳米工厂,该工厂可以修饰嵌入在DNA折纸平台上的目标DNA。该酶被编程为驻留在靶DNA的附近,与基于溶液的DNA修饰相比,可大大提高局部浓度。为了证明这一点,我们将DNA甲基转移酶M·TaqI固定在DNA折纸上的目标DNA旁边,并使用该酶通过辅因子类似物用生物素对目标DNA进行序列特异性修饰。链霉亲和素与生物素的结合被用作地形标记,以使用原子力显微镜成像技术跟踪该酶纳米工厂的机械循环。纳米工厂被证明是可回收的,并具有扩展到由DNA,RNA或蛋白质组成的简单到复杂分子控制的多酶,多底物操作系统的潜力。
更新日期:2021-01-20
中文翻译:

基于折纸的酶纳米工厂的DNA修饰和可视化
在过去的十年中,随着DNA折纸的出现,DNA纳米技术取得了巨大进步。使DNA折纸具有多种用途的功能是该领域的近期重点。在这里,我们构建了一个新型的DNA酶纳米工厂,该工厂可以修饰嵌入在DNA折纸平台上的目标DNA。该酶被编程为驻留在靶DNA的附近,与基于溶液的DNA修饰相比,可大大提高局部浓度。为了证明这一点,我们将DNA甲基转移酶M·TaqI固定在DNA折纸上的目标DNA旁边,并使用该酶通过辅因子类似物用生物素对目标DNA进行序列特异性修饰。链霉亲和素与生物素的结合被用作地形标记,以使用原子力显微镜成像技术跟踪该酶纳米工厂的机械循环。纳米工厂被证明是可回收的,并具有扩展到由DNA,RNA或蛋白质组成的简单到复杂分子控制的多酶,多底物操作系统的潜力。



























 京公网安备 11010802027423号
京公网安备 11010802027423号