当前位置:
X-MOL 学术
›
NMR Biomed.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Deep learning magnetic resonance spectroscopy fingerprints of brain tumours using quantum mechanically synthesised data
NMR in Biomedicine ( IF 2.9 ) Pub Date : 2021-01-14 , DOI: 10.1002/nbm.4479 Nikolaos Dikaios 1, 2
NMR in Biomedicine ( IF 2.9 ) Pub Date : 2021-01-14 , DOI: 10.1002/nbm.4479 Nikolaos Dikaios 1, 2
Affiliation
|
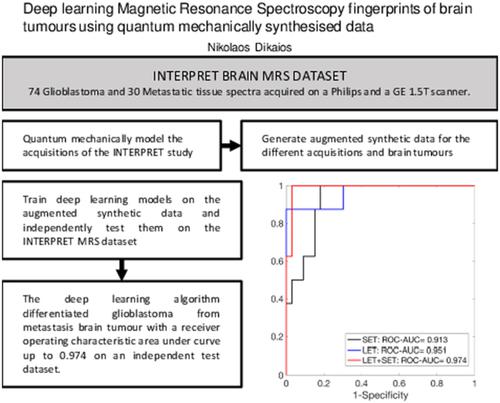
Metabolic fingerprints are valuable biomarkers for diseases that are associated with metabolic disorders. 1H magnetic resonance spectroscopy (MRS) is a unique noninvasive diagnostic tool that can depict the metabolic fingerprint based solely on the proton signal of different molecules present in the tissue. However, its performance is severely hindered by low SNR, field inhomogeneities and overlapping spectra of metabolites, which affect the quantification of metabolites. Consequently, MRS is rarely included in routine clinical protocols and has not been proven in multi‐institutional trials. This work proposes an alternative approach, where instead of quantifying metabolites’ concentration, deep learning (DL) is used to model the complex nonlinear relationship between diseases and their spectroscopic metabolic fingerprint (pattern). DL requires large training datasets, acquired (ideally) with the same protocol/scanner, which are very rarely available. To overcome this limitation, a novel method is proposed that can quantum mechanically synthesise MRS data for any scanner/acquisition protocol. The proposed methodology is applied to the challenging clinical problem of differentiating metastasis from glioblastoma brain tumours on data acquired across multiple institutions. DL algorithms were trained on the augmented synthetic spectra and tested on two independent datasets acquired by different scanners, achieving a receiver operating characteristic area under the curve of up to 0.96 and 0.97, respectively.
中文翻译:

使用量子力学合成数据的脑肿瘤深度学习磁共振波谱指纹图谱
代谢指纹是与代谢紊乱相关的疾病的宝贵生物标志物。1H 磁共振波谱 (MRS) 是一种独特的无创诊断工具,可以仅根据组织中存在的不同分子的质子信号来描绘代谢指纹。然而,其性能受到低信噪比、场不均匀性和代谢物光谱重叠的严重阻碍,影响了代谢物的定量。因此,MRS 很少包含在常规临床方案中,并且尚未在多机构试验中得到证实。这项工作提出了一种替代方法,该方法不是量化代谢物的浓度,而是使用深度学习 (DL) 来模拟疾病与其光谱代谢指纹(模式)之间复杂的非线性关系。DL 需要大型训练数据集,(理想情况下)使用相同的协议/扫描仪获取,而这些数据很少可用。为了克服这一限制,提出了一种新方法,该方法可以量子机械合成任何扫描仪/采集协议的 MRS 数据。所提出的方法适用于具有挑战性的临床问题,即根据跨多个机构获得的数据区分胶质母细胞瘤脑肿瘤的转移。DL 算法在增强的合成光谱上进行了训练,并在由不同扫描仪获取的两个独立数据集上进行了测试,分别实现了高达 0.96 和 0.97 的曲线下接收器操作特征区域。提出了一种新方法,可以为任何扫描仪/采集协议以量子机械方式合成 MRS 数据。所提出的方法适用于具有挑战性的临床问题,即根据跨多个机构获得的数据区分胶质母细胞瘤脑肿瘤的转移。DL 算法在增强的合成光谱上进行了训练,并在由不同扫描仪获取的两个独立数据集上进行了测试,分别实现了高达 0.96 和 0.97 的曲线下接收器操作特征区域。提出了一种新方法,可以为任何扫描仪/采集协议以量子机械方式合成 MRS 数据。所提出的方法适用于具有挑战性的临床问题,即根据跨多个机构获得的数据区分胶质母细胞瘤脑肿瘤的转移。DL 算法在增强的合成光谱上进行了训练,并在由不同扫描仪获取的两个独立数据集上进行了测试,分别实现了高达 0.96 和 0.97 的曲线下接收器操作特征区域。
更新日期:2021-03-07
中文翻译:

使用量子力学合成数据的脑肿瘤深度学习磁共振波谱指纹图谱
代谢指纹是与代谢紊乱相关的疾病的宝贵生物标志物。1H 磁共振波谱 (MRS) 是一种独特的无创诊断工具,可以仅根据组织中存在的不同分子的质子信号来描绘代谢指纹。然而,其性能受到低信噪比、场不均匀性和代谢物光谱重叠的严重阻碍,影响了代谢物的定量。因此,MRS 很少包含在常规临床方案中,并且尚未在多机构试验中得到证实。这项工作提出了一种替代方法,该方法不是量化代谢物的浓度,而是使用深度学习 (DL) 来模拟疾病与其光谱代谢指纹(模式)之间复杂的非线性关系。DL 需要大型训练数据集,(理想情况下)使用相同的协议/扫描仪获取,而这些数据很少可用。为了克服这一限制,提出了一种新方法,该方法可以量子机械合成任何扫描仪/采集协议的 MRS 数据。所提出的方法适用于具有挑战性的临床问题,即根据跨多个机构获得的数据区分胶质母细胞瘤脑肿瘤的转移。DL 算法在增强的合成光谱上进行了训练,并在由不同扫描仪获取的两个独立数据集上进行了测试,分别实现了高达 0.96 和 0.97 的曲线下接收器操作特征区域。提出了一种新方法,可以为任何扫描仪/采集协议以量子机械方式合成 MRS 数据。所提出的方法适用于具有挑战性的临床问题,即根据跨多个机构获得的数据区分胶质母细胞瘤脑肿瘤的转移。DL 算法在增强的合成光谱上进行了训练,并在由不同扫描仪获取的两个独立数据集上进行了测试,分别实现了高达 0.96 和 0.97 的曲线下接收器操作特征区域。提出了一种新方法,可以为任何扫描仪/采集协议以量子机械方式合成 MRS 数据。所提出的方法适用于具有挑战性的临床问题,即根据跨多个机构获得的数据区分胶质母细胞瘤脑肿瘤的转移。DL 算法在增强的合成光谱上进行了训练,并在由不同扫描仪获取的两个独立数据集上进行了测试,分别实现了高达 0.96 和 0.97 的曲线下接收器操作特征区域。



























 京公网安备 11010802027423号
京公网安备 11010802027423号