Journal of Proteomics ( IF 3.3 ) Pub Date : 2021-01-14 , DOI: 10.1016/j.jprot.2021.104114 Max Gilbert 1 , Zhi Li 1 , Xu Na Wu 2 , Leander Rohr 3 , Sven Gombos 1 , Klaus Harter 3 , Waltraud X Schulze 1
|
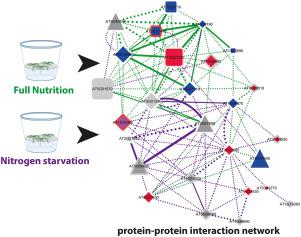
Plants must rapidly adapt to changes in nutrient conditions. Especially adaptations to changing nitrogen environments are very complex involving also major adjustments on the protein level. Here, we used a size-exclusion chromatography-coupled to mass spectrometry approach to study the dynamics of protein-protein interactions induced by transition from full nutrition to nitrogen starvation. Comparison of interaction networks established for each nutrient condition revealed a large overlap of proteins which were part of the protein-protein interaction network, but that same set of proteins underwent different interactions at each treatment. Network topology parameter betweenness centrality (BC) was found to best reflect the relevance of individual proteins in the information flow within each network. Changes in BC for individual proteins may therefore indicate their involvement in the cellular adjustments to the new condition. Based on this analysis, a set of proteins was identified showing high nitrogen-dependent changes in their BC values: The receptor kinase AT5G49770, co-receptor QSK1, and proton-ATPase AHA2. Mutants of those proteins showed a nitrate-dependent root growth phenotype. Individual interactions within the reconstructed network were tested using FRET-FLIM technology. Taken together, we present a systematic strategy comparing dynamic changes in protein-protein interaction networks based on their network parameters to identify regulatory nodes.
Significance
Protein-protein interactions are known to be important in cellular signaling events, but the dynamic changes in interaction networks induced by external stimuli are still rarely studied. We systematically analyzed how changes in the nutrient environment induced a rewiring of protein-protein interactions in roots. We observed small changes in overall protein abundances, but instead a rewiring of pairwise protein-protein interactions. Betweenness centrality was found to be the optimal network topology parameter to identify protein candidates with high relevance to the information flow in the (dynamic) network. Predicted interactions of those relevant nodes were confirmed in FLIM/FRET experiments and in phenotypic analysis. The network approach described here may be a useful application in dynamic network analysis more generally.
中文翻译:

蛋白质-蛋白质相互作用网络中基于路径的中心度测量方法的比较揭示了在适应变化的氮环境期间具有表型相关性的蛋白质
植物必须迅速适应营养条件的变化。特别是对于不断变化的氮环境的适应非常复杂,其中还涉及蛋白质水平的重大调整。在这里,我们使用了体积排阻色谱-质谱联用的方法,研究了从完全营养过渡到氮饥饿引起的蛋白质-蛋白质相互作用的动力学。针对每种营养条件建立的相互作用网络的比较显示,蛋白质之间存在很大的重叠,这是蛋白质-蛋白质相互作用网络的一部分,但是同一组蛋白质在每次处理中经历了不同的相互作用。发现网络拓扑参数之间的中心度(BC)可以最好地反映每个网络内信息流中各个蛋白质的相关性。因此,单个蛋白质的BC变化可能表明它们参与了对新情况的细胞调节。基于此分析,确定了一组蛋白质,这些蛋白质在其BC值中显示出氮依赖性高的变化:受体激酶AT5G49770,共受体QSK1和质子-ATPase AHA2。这些蛋白质的突变体显示出硝酸盐依赖性根生长表型。使用FRET-FLIM技术测试了重构网络中的各个交互。综上所述,我们提出了一种系统策略,比较基于蛋白质网络相互作用参数的蛋白质相互作用相互作用网络的动态变化,以识别调控节点。鉴定出一组蛋白质,这些蛋白质在其BC值中显示出高氮依赖性变化:受体激酶AT5G49770,共受体QSK1和质子-ATPase AHA2。这些蛋白质的突变体显示出硝酸盐依赖性根生长表型。使用FRET-FLIM技术测试了重建网络中的各个交互。综上所述,我们提出了一种系统策略,比较基于蛋白质网络相互作用参数的蛋白质相互作用相互作用网络的动态变化,以识别调控节点。鉴定出一组蛋白质,这些蛋白质在其BC值中显示出高氮依赖性变化:受体激酶AT5G49770,共受体QSK1和质子-ATPase AHA2。这些蛋白质的突变体显示出硝酸盐依赖性根生长表型。使用FRET-FLIM技术测试了重构网络中的各个交互。综上所述,我们提出了一种系统策略,比较基于蛋白质网络相互作用参数的蛋白质相互作用相互作用网络的动态变化,以识别调控节点。
意义
蛋白质间相互作用在细胞信号事件中是重要的,但是由外部刺激引起的相互作用网络的动态变化仍然很少被研究。我们系统地分析了营养环境的变化如何导致根部蛋白质间相互作用的重新结合。我们观察到总蛋白质丰度的微小变化,但是成对蛋白质-蛋白质相互作用的重新布线。发现中间性中心是确定与(动态)网络中的信息流高度相关的蛋白质候选物的最佳网络拓扑参数。在FLIM / FRET实验和表型分析中证实了这些相关节点的预测相互作用。一般而言,此处描述的网络方法在动态网络分析中可能是有用的应用程序。


























 京公网安备 11010802027423号
京公网安备 11010802027423号