当前位置:
X-MOL 学术
›
Mol. Omics
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Comparison between instrumental variable and mediation-based methods for reconstructing causal gene networks in yeast
Molecular Omics ( IF 2.9 ) Pub Date : 2020-12-17 , DOI: 10.1039/d0mo00140f Adriaan-Alexander Ludl 1 , Tom Michoel
Molecular Omics ( IF 2.9 ) Pub Date : 2020-12-17 , DOI: 10.1039/d0mo00140f Adriaan-Alexander Ludl 1 , Tom Michoel
Affiliation
|
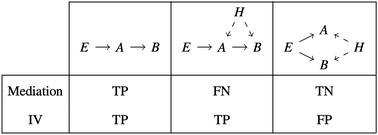
Causal gene networks model the flow of information within a cell. Reconstructing causal networks from omics data is challenging because correlation does not imply causation. When genomics and transcriptomics data from a segregating population are combined, genomic variants can be used to orient the direction of causality between gene expression traits. Instrumental variable methods use a local expression quantitative trait locus (eQTL) as a randomized instrument for a gene's expression level, and assign target genes based on distal eQTL associations. Mediation-based methods additionally require that distal eQTL associations are mediated by the source gene. A detailed comparison between these methods has not yet been conducted, due to the lack of a standardized implementation of different methods, the limited sample size of most multi-omics datasets, and the absence of ground-truth networks for most organisms. Here we used Findr, a software package providing uniform implementations of instrumental variable, mediation, and coexpression-based methods, a recent dataset of 1012 segregants from a cross between two budding yeast strains, and the YEASTRACT database of known transcriptional interactions to compare causal gene network inference methods. We found that causal inference methods result in a significant overlap with the ground-truth, whereas coexpression did not perform better than random. A subsampling analysis revealed that the performance of mediation saturates at large sample sizes, due to a loss of sensitivity when residual correlations become significant. Instrumental variable methods on the other hand contain false positive predictions, due to genomic linkage between eQTL instruments. Instrumental variable and mediation-based methods also have complementary roles for identifying causal genes underlying transcriptional hotspots. Instrumental variable methods correctly predicted STB5 targets for a hotspot centred on the transcription factor STB5, whereas mediation failed due to Stb5p auto-regulating its own expression. Mediation suggests a new candidate gene, DNM1, for a hotspot on Chr XII, whereas instrumental variable methods could not distinguish between multiple genes located within the hotspot. In conclusion, causal inference from genomics and transcriptomics data is a powerful approach for reconstructing causal gene networks, which could be further improved by the development of methods to control for residual correlations in mediation analyses, and for genomic linkage and pleiotropic effects from transcriptional hotspots in instrumental variable analyses.
中文翻译:

用于重建酵母因果基因网络的工具变量和基于中介的方法的比较
因果基因网络对细胞内的信息流进行建模。从组学数据重建因果网络具有挑战性,因为相关性并不意味着因果关系。当来自分离群体的基因组学和转录组学数据结合起来时,基因组变异可用于确定基因表达特征之间因果关系的方向。工具变量方法使用局部表达数量性状基因座 (eQTL) 作为基因表达水平的随机工具,并根据远端 eQTL 关联分配目标基因。基于中介的方法还要求远端 eQTL 关联由源基因介导。由于缺乏不同方法的标准化实施,尚未对这些方法进行详细比较,大多数多组学数据集的样本量有限,并且大多数生物体缺乏真实网络。在这里,我们使用了 Findr,这是一个软件包,提供了工具变量、中介和基于共表达的方法的统一实现,最近的数据集来自两个芽殖酵母菌株之间的 1012 个分离子,以及 YEASTRACT已知转录相互作用数据库,用于比较因果基因网络推断方法。我们发现因果推断方法导致与真实情况的显着重叠,而共表达的表现并不比随机更好。子抽样分析表明,由于当残差相关性变得显着时敏感性损失,中介的性能在大样本量下饱和。另一方面,由于 eQTL 仪器之间的基因组联系,仪器变量方法包含假阳性预测。工具变量和基于中介的方法在识别转录热点下的因果基因方面也具有互补作用。工具变量方法正确预测STB5热点的目标以转录因子STB5为中心,而由于 Stb5p 自动调节其自身表达,介导失败。中介建议新的候选基因DNM1用于 Chr XII 上的热点,而工具变量方法无法区分位于热点内的多个基因。总之,基因组学和转录组学数据的因果推断是重建因果基因网络的有力方法,可以通过开发方法来控制中介分析中的残余相关性,以及控制来自转录热点的基因组连锁和多效性效应的方法来进一步改进。工具变量分析。
更新日期:2021-01-13
中文翻译:

用于重建酵母因果基因网络的工具变量和基于中介的方法的比较
因果基因网络对细胞内的信息流进行建模。从组学数据重建因果网络具有挑战性,因为相关性并不意味着因果关系。当来自分离群体的基因组学和转录组学数据结合起来时,基因组变异可用于确定基因表达特征之间因果关系的方向。工具变量方法使用局部表达数量性状基因座 (eQTL) 作为基因表达水平的随机工具,并根据远端 eQTL 关联分配目标基因。基于中介的方法还要求远端 eQTL 关联由源基因介导。由于缺乏不同方法的标准化实施,尚未对这些方法进行详细比较,大多数多组学数据集的样本量有限,并且大多数生物体缺乏真实网络。在这里,我们使用了 Findr,这是一个软件包,提供了工具变量、中介和基于共表达的方法的统一实现,最近的数据集来自两个芽殖酵母菌株之间的 1012 个分离子,以及 YEASTRACT已知转录相互作用数据库,用于比较因果基因网络推断方法。我们发现因果推断方法导致与真实情况的显着重叠,而共表达的表现并不比随机更好。子抽样分析表明,由于当残差相关性变得显着时敏感性损失,中介的性能在大样本量下饱和。另一方面,由于 eQTL 仪器之间的基因组联系,仪器变量方法包含假阳性预测。工具变量和基于中介的方法在识别转录热点下的因果基因方面也具有互补作用。工具变量方法正确预测STB5热点的目标以转录因子STB5为中心,而由于 Stb5p 自动调节其自身表达,介导失败。中介建议新的候选基因DNM1用于 Chr XII 上的热点,而工具变量方法无法区分位于热点内的多个基因。总之,基因组学和转录组学数据的因果推断是重建因果基因网络的有力方法,可以通过开发方法来控制中介分析中的残余相关性,以及控制来自转录热点的基因组连锁和多效性效应的方法来进一步改进。工具变量分析。



























 京公网安备 11010802027423号
京公网安备 11010802027423号