当前位置:
X-MOL 学术
›
Acta Cryst. D
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Ab initio structure solution of proteins at atomic resolution using charge‐flipping techniques and cloud computing
Acta Crystallographica Section D ( IF 2.2 ) Pub Date : 2021-01-06 , DOI: 10.1107/s2059798320015090 Alan A Coelho 1
Acta Crystallographica Section D ( IF 2.2 ) Pub Date : 2021-01-06 , DOI: 10.1107/s2059798320015090 Alan A Coelho 1
Affiliation
|
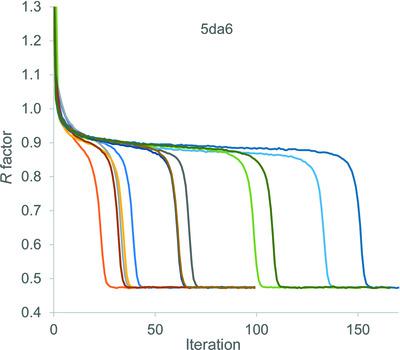
Large protein structures at atomic resolution can be solved in minutes using charge‐flipping techniques operating on hundreds of virtual machines (computers) on the Amazon Web Services cloud‐computing platform driven by the computer programs TOPAS or TOPAS‐Academic at a small financial cost. The speed of operation has allowed charge‐flipping techniques to be investigated and modified, leading to two strategies that can solve a large range of difficult protein structures at atomic resolution. Techniques include the use of space‐group symmetry restraints on the electron density as well as increasing the intensity of a randomly chosen high‐intensity electron‐density peak. It is also shown that the use of symmetry restraints increases the chance of finding a solution for low‐resolution data. Finally, a flipping strategy that negates `uranium atom solutions' has been developed for structures that exhibit such solutions during charge flipping.
中文翻译:

使用电荷翻转技术和云计算在原子分辨率下从头算蛋白质结构解
在由计算机程序TOPAS或TOPAS-Academic驱动的 Amazon Web Services 云计算平台上的数百台虚拟机(计算机)上运行的电荷翻转技术可以在几分钟内解决原子分辨率的大型蛋白质结构以很小的财务成本。操作的速度允许研究和修改电荷翻转技术,从而产生两种可以在原子分辨率下解决大范围困难蛋白质结构的策略。技术包括对电子密度使用空间群对称约束以及增加随机选择的高强度电子密度峰的强度。还表明,对称约束的使用增加了为低分辨率数据找到解决方案的机会。最后,已经为在电荷翻转过程中表现出这种解决方案的结构开发了一种否定“铀原子溶液”的翻转策略。
更新日期:2021-01-06
中文翻译:

使用电荷翻转技术和云计算在原子分辨率下从头算蛋白质结构解
在由计算机程序TOPAS或TOPAS-Academic驱动的 Amazon Web Services 云计算平台上的数百台虚拟机(计算机)上运行的电荷翻转技术可以在几分钟内解决原子分辨率的大型蛋白质结构以很小的财务成本。操作的速度允许研究和修改电荷翻转技术,从而产生两种可以在原子分辨率下解决大范围困难蛋白质结构的策略。技术包括对电子密度使用空间群对称约束以及增加随机选择的高强度电子密度峰的强度。还表明,对称约束的使用增加了为低分辨率数据找到解决方案的机会。最后,已经为在电荷翻转过程中表现出这种解决方案的结构开发了一种否定“铀原子溶液”的翻转策略。


























 京公网安备 11010802027423号
京公网安备 11010802027423号