当前位置:
X-MOL 学术
›
FEBS Open Bio
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Development and validation of a four-lipid metabolism gene signature for diagnosis of pancreatic cancer
FEBS Open Bio ( IF 2.6 ) Pub Date : 2021-01-01 , DOI: 10.1002/2211-5463.13074 Yanrong Ye 1, 2 , Zhe Chen 1 , Yun Shen 1 , Yan Qin 1 , Hao Wang 3
FEBS Open Bio ( IF 2.6 ) Pub Date : 2021-01-01 , DOI: 10.1002/2211-5463.13074 Yanrong Ye 1, 2 , Zhe Chen 1 , Yun Shen 1 , Yan Qin 1 , Hao Wang 3
Affiliation
|
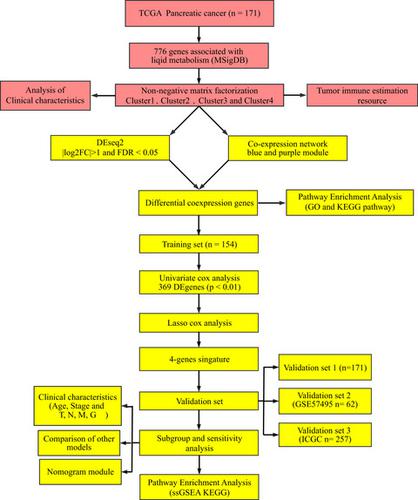
Abnormal lipid metabolism is closely related to the malignant biological behavior of tumor cells. Such abnormal lipid metabolism provides energy for rapid proliferation, and certain genes related to lipid metabolism encode important components of tumor signaling pathways. In this study, we analyzed pancreatic cancer datasets from The Cancer Genome Atlas and searched for prognostic genes related to lipid metabolism in the Molecular Signature Database. A risk score model was built and verified using the GSE57495 dataset and International Cancer Genome Consortium dataset. Four molecular subtypes and 4249 differentially expressed genes (DEGs) were identified. The DEGs obtained by Weighted Gene Coexpression Network Construction analysis were intersected with 4249 DEGs to obtain a total of 1340 DEGs. The final prognosis model included CA8, CEP55, GNB3 and SGSM2, and these had a significant effect on overall survival. The area under the curve at 1, 3 and 5 years was 0.72, 0.79 and 0.87, respectively. These same results were obtained using the validation cohort. Survival analysis data showed that the model could stratify the prognosis of patients with different clinical characteristics, and the model has clinical independence. Functional analysis indicated that the model is associated with multiple cancer-related pathways. Compared with published models, our model has a higher C-index and greater risk value. In summary, this four-gene signature is an independent risk factor for pancreatic cancer survival and may be an effective prognostic indicator.
中文翻译:

用于诊断胰腺癌的四脂代谢基因特征的开发和验证
脂质代谢异常与肿瘤细胞的恶性生物学行为密切相关。这种异常的脂质代谢为快速增殖提供了能量,某些与脂质代谢相关的基因编码了肿瘤信号通路的重要成分。在这项研究中,我们分析了来自癌症基因组图谱的胰腺癌数据集,并在分子特征数据库中搜索了与脂质代谢相关的预后基因。使用 GSE57495 数据集和国际癌症基因组联盟数据集建立并验证了风险评分模型。鉴定了四种分子亚型和4249个差异表达基因(DEG)。将加权基因共表达网络构建分析得到的DEGs与4249个DEGs相交共得到1340个DEGs。最终的预后模型包括CA8、CEP55、GNB3和SGSM2,这些对总生存率有显着影响。1、3 和 5 年的曲线下面积分别为 0.72、0.79 和 0.87。这些相同的结果是使用验证队列获得的。生存分析数据表明,该模型可以对不同临床特征患者的预后进行分层,模型具有临床独立性。功能分析表明,该模型与多种癌症相关途径相关。与已发表的模型相比,我们的模型具有更高的 C 指数和更大的风险值。总之,这种四基因特征是胰腺癌存活的独立危险因素,可能是有效的预后指标。
更新日期:2021-01-01
中文翻译:

用于诊断胰腺癌的四脂代谢基因特征的开发和验证
脂质代谢异常与肿瘤细胞的恶性生物学行为密切相关。这种异常的脂质代谢为快速增殖提供了能量,某些与脂质代谢相关的基因编码了肿瘤信号通路的重要成分。在这项研究中,我们分析了来自癌症基因组图谱的胰腺癌数据集,并在分子特征数据库中搜索了与脂质代谢相关的预后基因。使用 GSE57495 数据集和国际癌症基因组联盟数据集建立并验证了风险评分模型。鉴定了四种分子亚型和4249个差异表达基因(DEG)。将加权基因共表达网络构建分析得到的DEGs与4249个DEGs相交共得到1340个DEGs。最终的预后模型包括CA8、CEP55、GNB3和SGSM2,这些对总生存率有显着影响。1、3 和 5 年的曲线下面积分别为 0.72、0.79 和 0.87。这些相同的结果是使用验证队列获得的。生存分析数据表明,该模型可以对不同临床特征患者的预后进行分层,模型具有临床独立性。功能分析表明,该模型与多种癌症相关途径相关。与已发表的模型相比,我们的模型具有更高的 C 指数和更大的风险值。总之,这种四基因特征是胰腺癌存活的独立危险因素,可能是有效的预后指标。


























 京公网安备 11010802027423号
京公网安备 11010802027423号