当前位置:
X-MOL 学术
›
Microbiologyopen
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Temporal transcriptomes of a marine cyanopodovirus and its Synechococcus host during infection
MicrobiologyOpen ( IF 3.4 ) Pub Date : 2020-12-30 , DOI: 10.1002/mbo3.1150 Sijun Huang 1 , Yingting Sun 1 , Si Zhang 1 , Lijuan Long 1
MicrobiologyOpen ( IF 3.4 ) Pub Date : 2020-12-30 , DOI: 10.1002/mbo3.1150 Sijun Huang 1 , Yingting Sun 1 , Si Zhang 1 , Lijuan Long 1
Affiliation
|
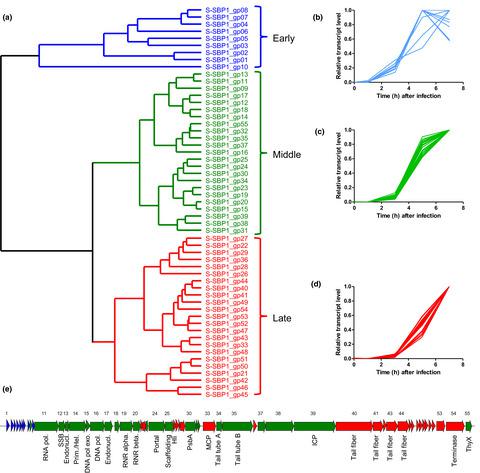
Marine picocyanobacteria belonging to genera Synechococcus and Prochlorococcus are genetically diverged and distributed into distinct biogeographical patterns, and both are infected by genetically closely related cyanopodoviruses. Previous studies have not fully explored whether the two virus–host systems share similar gene expression patterns during infection. Whole‐genome expression dynamics of T7‐like cyanopodovirus P‐SSP7 and its host Prochlorococcus strain MED4 have already been reported. Here, we conducted genomic and transcriptomic analyses on T7‐like cyanopodovirus S‐SBP1 during its infection on Synechococcus strain WH7803. S‐SBP1 has a latent period of 8 h and phage DNA production of 30 copies per cell. In terms of whole‐genome phylogenetic relationships and average nucleotide identity, S‐SBP1 was most similar to cyanopodovirus S‐RIP2, which also infects Synechococcus WH7803. Three hypervariable genomic islands were identified when comparing the genomes of S‐SBP1 and S‐RIP2. Single nucleotide variants were also observed in three S‐SBP1 genes, which were located within the island regions. Based on RNA‐seq analysis, S‐SBP1 genes clustered into three temporal expression classes, whose gene content was similar to that of P‐SSP7. Thirty‐two host genes were upregulated during phage infection, including those involved in carbon metabolism, ribosome components, and stress response. These upregulated genes were similar to those upregulated by Prochlorococcus MED4 in response to infection by P‐SSP7. Our study demonstrates a programmed temporal expression pattern of cyanopodoviruses and hosts during infection.
中文翻译:

海洋蓝藻病毒及其聚球藻宿主在感染过程中的时间转录组
属于Synechococcus和Prochlorococcus属的海洋微微蓝细菌在遗传上分化并分布成不同的生物地理模式,并且两者都被遗传上密切相关的蓝藻病毒感染。以前的研究尚未充分探讨两种病毒宿主系统在感染期间是否具有相似的基因表达模式。已经报道了 T7 样蓝藻病毒 P-SSP7 及其宿主原绿球藻菌株 MED4 的全基因组表达动态。在这里,我们对 T7 样蓝藻病毒 S-SBP1 在其感染聚球藻的过程中进行了基因组和转录组学分析菌株 WH7803。S-SBP1 的潜伏期为 8 小时,每个细胞产生 30 个噬菌体 DNA 拷贝。在全基因组系统发育关系和平均核苷酸同一性方面,S-SBP1 与同样感染聚球藻属的蓝斑病病毒 S-RIP2 最相似WH7803。在比较 S-SBP1 和 S-RIP2 的基因组时,确定了三个高变基因组岛。在位于岛屿区域内的三个 S-SBP1 基因中也观察到了单核苷酸变异。基于 RNA-seq 分析,S-SBP1 基因聚集成三个时间表达类别,其基因含量与 P-SSP7 相似。32 个宿主基因在噬菌体感染期间被上调,包括那些参与碳代谢、核糖体成分和应激反应的基因。这些上调基因与Prochlorococcus MED4 响应 P-SSP7 感染上调的基因相似。我们的研究证明了在感染过程中蓝藻病毒和宿主的程序化时间表达模式。
更新日期:2021-02-16
中文翻译:

海洋蓝藻病毒及其聚球藻宿主在感染过程中的时间转录组
属于Synechococcus和Prochlorococcus属的海洋微微蓝细菌在遗传上分化并分布成不同的生物地理模式,并且两者都被遗传上密切相关的蓝藻病毒感染。以前的研究尚未充分探讨两种病毒宿主系统在感染期间是否具有相似的基因表达模式。已经报道了 T7 样蓝藻病毒 P-SSP7 及其宿主原绿球藻菌株 MED4 的全基因组表达动态。在这里,我们对 T7 样蓝藻病毒 S-SBP1 在其感染聚球藻的过程中进行了基因组和转录组学分析菌株 WH7803。S-SBP1 的潜伏期为 8 小时,每个细胞产生 30 个噬菌体 DNA 拷贝。在全基因组系统发育关系和平均核苷酸同一性方面,S-SBP1 与同样感染聚球藻属的蓝斑病病毒 S-RIP2 最相似WH7803。在比较 S-SBP1 和 S-RIP2 的基因组时,确定了三个高变基因组岛。在位于岛屿区域内的三个 S-SBP1 基因中也观察到了单核苷酸变异。基于 RNA-seq 分析,S-SBP1 基因聚集成三个时间表达类别,其基因含量与 P-SSP7 相似。32 个宿主基因在噬菌体感染期间被上调,包括那些参与碳代谢、核糖体成分和应激反应的基因。这些上调基因与Prochlorococcus MED4 响应 P-SSP7 感染上调的基因相似。我们的研究证明了在感染过程中蓝藻病毒和宿主的程序化时间表达模式。



























 京公网安备 11010802027423号
京公网安备 11010802027423号